Does human papillomavirus cause human colorectal cancer? Applying Bradford Hill criteria postulates
Muhammad Usman, Yasir Hameed and Mukhtiar Ahmad
Department of Biochemistry and Biotechnology, The Islamia University of Bahawalpur, Bahawalpur, Pakistan
Abstract
The role of human papillomavirus (HPV) in human colorectal cancer (CRC) has already been widely investigated worldwide with conflicting results. Although researchers have tried to establish the link between HPV and CRC through a statistical meta-analysis of the previous studies associating HPV with CRC, they failed to establish a more reliable link due to the shortcomings of the statistical meta-analysis. In the present study, we identified population-wide studies relating HPV with CRC through the PubMed search engine. Then, we examined the available data of HPV prevalence in CRC and normal/benign samples and applied the postulates of Bradford Hill criteria on the available evidence to investigate the association between HPV and CRC. The Bradford Hill criteria are very old, reliable and widely accepted for establishing a link between the cause and disease. In addition, to further enhance the reliability of the outcomes, we have also evaluated the methodologies of the previous studies to address the possibility of false-negative and false-positive results. After a careful evaluation of the extracted data against the postulates of Bradford Hill criteria, it was observed that none of the studies fulfil all the major postulates of Bradford Hill criteria for causation including temporality, consistency, biological gradient, experiment, coherence, specificity and analogy. Hence, no causal relationship has been suggested between HPV and CRC patients of the any included population. The results failed to prove the causal relationship between HPV and CRC and suggested HPV as a coparticipant in the pathogenesis of CRC.
Keywords: human papillomavirus (HPV), colorectal cancer (CRC), PubMed, Bradford Hill criteria, pathogenesis
Correspondence to: Yasir Hameed
Email: Yasirhameed2011@gmail.com
Published: 21/09/2020
Received: 14/04/2020
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Colorectal cancer (CRC), also known as large bowel cancer, is a cancer of the colon, rectum and appendix [1]. In general, it develops gradually within 10–15 years. Patients with ulcerative colitis are at a higher risk of developing CRC compared to the general population [2]. Globally, CRC is the third most commonly diagnosed cancer in men and the second most frequently reported cancer in women [3]. In the United States, it is estimated that 147,950 new cases and 5200 CRC related deaths will occur in 2020 [4]. CRC is more prevalent in developed countries accounting for 63% of cases of all the cancer cases. The high-risk hotspots of CRC include Europe, Australia and North America. In these areas, CRC is more prevalent amongst urban residents than the rural residents [5].
More than 100 subtypes of HPV have been reported in the medical literature until now, of which 40 subtypes are known to infect the genital epithelial cells [6]. At least 15 HPV subtypes including HPV 16, 18, 31, 35, 39, 45, 51, 52, 56, 59, 66, 68, 69, 73 and 82 are classified as high-risk subtypes due to the significant association with genital tract and non-genital tract malignancy [7].
It has already been acknowledged earlier that two HPV viral oncogenes (E6 and E7) contribute majorly to the development of HPV-induced CRC. Oncoprotein E7 inactivates hypophosphorylated retinoblastoma protein (pRB) by tightly binding with it, and then, the inactivation of pRB eventually results in the upregulation of p16INK4A. P16INK4A is a tumour suppressor protein that is involved in inhibiting the cyclin-dependent kinases 6 and 4, which regulates the G1 checkpoint of the cell cycle. The role of oncoprotein E6 is well recognised for functionally inactivating p53 protein which is a major regulator of the cell cycle [8]. In earlier studies, a reported inverse correlation between p53 mutations and HPV infection suggested that HPV targets and inactivates the p53, and in turn, the inactivated p53 significantly contributes to the development and progression of CRC by dysregulating various important pathways [9].
The first-ever study documenting the presence of HPV was conducted in 1990 by Kirgan et al [10] in the Czechoslovakia population. In that study, they detected the presence of HPV in a total of 73 paraffin-embedded CRC samples and 30 normal colon mucosal tissue samples using the Southern blotting technique. The results of their study revealed the presence of HPV in approximately 82% and 23% of CRC and normal controls, respectively.
Since then, numerous studies have been carried out worldwide for detecting HPV in CRC, and their outcomes were contradictory because they detected HPV in varying frequencies in different populations, i.e., from 0% [11, 12] to 100% [13].
In general, a statistical meta-analysis is usually preferred when establishing a correlation between the virus and disease as compared to the single study. This choice is based on the multiple advantages of the meta-analysis such as increased number of objects, greater diversity amongst the objects and conclusion with a high level of evidence over the individual single study, which has disadvantages like a small cohort of patients and conclusions with a low level of evidence. By keeping in view the inconsistencies in the HPV detection ratios in worldwide published studies, recently, researchers have analysed the previously published studies by the means of statistical meta-analysis to yield more useful pieces of information.
Previously, a statistical meta-analysis was performed to find out the causal relationship between HPV and CRC by Damin et al [14] of the available literature on HPV detection in CRC by searching various authentic research engines including Medline (PubMed), Embase (OVID) and Web of Science. They obtained more than 18 studies from different populations such as Europe, Asian and American. The results of their meta-analysis revealed that HPV infection significantly increased the risk of CRC development.
Similarly, Baandrup et al [15] performed another meta-analysis of the available literature reporting the association of HPV with CRC through various authentic research engines such as PubMed and Web of Science. They analysed 37 studies from American, Asia and Middle East populations. In their conclusion, they also reported a significant association between HPV and CRC.
Although evaluating the results of the previous studies documenting the role of HPV in the development of CRC through statistical meta-analysis was a better choice than generalising the results of an individual study, we did not consider statistical meta-analysis reliable to establish a causal relationship between HPV and the CRC development because of some serious limitations such as its inability to analyse the methodologies of the previous studies, so there is no way to evaluate the possibility of false-negative and false-positive results nor does statistical meta-analysis provide any information regarding the effect of heterogeneity-specific nature of the understudied populations on HPV detection. In addition, statistical meta-analysis results in publication biasness, where meta-analysis does not select studies with no results even though they contain valuable information.
By looking at the discrepancy in the outcomes of the previously published studies and significant shortcomings of the statistical meta-analysis, we performed the population-wide valuation of the results of the previous studies using the Bradford Hill criteria. These criteria are widely used and accepted worldwide over many years for establishing a causal relationship between a presumed cause and an observed effect on public health research [16].
In the course of evaluation, we analysed whether or not these studies fulfil all the postulates of Bradford Hill criteria to declare a causal relationship between HPV and CRC. In addition, we also evaluated the methodologies used by the previous studies to address the possibility of false-positive and false-negative results for better outcomes. The outcomes of the present study will help to establish a more reliable population-wide causal relationship between HPV and CRC and determine the more appropriate treatment strategies for CRC patients.
Methodology
In the present study, we implemented a two-phase methodology (Figure 1).
Literature search
All the relevant articles associating HPV with CRC were identified through the PubMed search database using the keywords: ‘Colorectal Cancer’ and ‘Human Papillomavirus’. We also defined ‘Papillomaviridae’ and ‘Colorectal neoplasia’ as medical subject headings (MeSH) terms. MeSH terms and keywords were combined during the search process. All the literature works were searched available until March 2020, with the ‘Original Article’ filter. In total, 1,363 original articles were identified through the PubMed search engine.
Relevant data extraction
From 1363 original articles, 59 relevant articles were identified having the desired information by initially reading the title, abstract and then complete article. Furthermore, a comprehensive table was constructed having all the required information from the selected relevant studies.
Evaluation of the results using the postulates of Bradford Hill criteria
Based on the extracted data, all the identified studies were carefully evaluated using the following postulates:
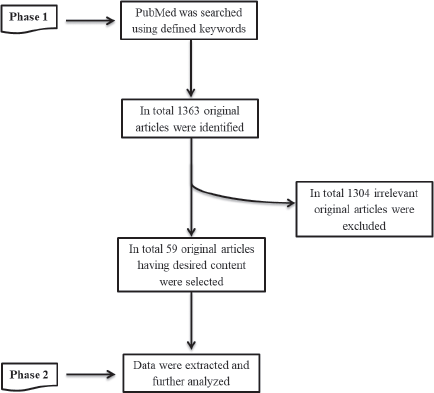
Figure 1. Overview of the methodology implemented during the present study.
(1) Strength: Larger the association, more probability of the causal relationship, (2) temporality: cause must lead to the induction of an effect. If the delay is expected between the cause and effect, then the effect has to occur after the delay, (3) consistency: different studies conducted by different researchers at different places with different sample sizes and reporting the similar results increase the chances of the causal relationship between the cause and effect, (4) plausibility: there should be plausible mechanism between the cause and effect, (5) biological gradient: greater response is produced by the causative agent in response to the greater exposure. However, in some cases, the effect can be triggered by the mere presence of the factor, whereas, in other cases, greater exposure can lead to lower effect as well, (6) experiment: the relationship between the cause and effect should be explained by the experiments, and the experiment should result in the reduction of effect when the causative agent is removed, (7) coherence: causal relationship should not conflict with already known literature about the disease or exposure, (8) specificity: causality is more likely if the effect has only one cause and (9) analogy: previous evidence of the association between the cause and effect should support the current statement for the causal relationship.
The assessment of each postulate was qualitative/descriptive, as there was an element of subjectivity in applying quantitative scoring. Evidence collected for each postulate is presented in Table 1 and the results section with a final judgment as to whether or not the viewpoint was fulfilled.
Results
In total, 59 relevant original articles (Table 1; Figure 2) were found on PubMed which investigated the association of HPV with CRC in 24 different populations. Table 1 shows all these articles and contains essential information extracted from them including the details of the studied population, techniques used for HPV detection, name of the target gene, the identified strains of HPV, the most prevalent identified stain and number (No) of screened samples (normal, benign and diseased) with their respective identified population-wide positivity ratios.
Of all the 59 studies, in total, only 43 studies [12, 17–58] were the case–control studies, in which normal, benign and CRC samples were screened, whereas others were not.
The positivity ratio of HPV detection in CRC samples was varied population wise from 0% [11, 12, 17, 23, 26, 32, 41, 43, 45, 46, 50, 59] to 100% [13, 54] in all the 59 studies, whereas the positivity ratio of HPV detection in normal and adjacent/benign samples was varied from 0% [21, 22, 29, 30, 43, 49] to 84% (28) and 0% [17, 22, 26, 41, 42, 45, 46, 50, 52, 57] to 69.56% [54], respectively.
The results obtained after careful evaluation of the extracted data through the Bradford Hill criteria postulates showed that all the identified studies from various populations do not fulfil the major postulates including temporality, consistency, biological gradient, experiment, coherence, specificity and analogy. Hence, no causal relationship has been suggested between HPV and CRC patients of any included population.
Polymerase chain reaction (PCR) technique was employed by most of the studies [8, 10, 11, 13, 17-35, 37–49, 52, 54, 55, 57–91] to detect the presence of HPV in the normal, adjacent/benign and diseased samples using L1, E6 and E7 gene-specific primers which specifically target [6, 11, 16, 18, 31, 33, 35, 39, 40, 42, 45, 51–59] the subtypes of HPV, and from them, in addition, few studies also employed the second techniques including immunohistochemistry [67] and Southern blotting [13, 28] to validate their PCR results.
Few studies also used immunohistochemistry [31, 36, 51, 56], gene chip technology [53], Southern blotting [50] and in situ hybridisation [12] for the detection of HPV, and they did not validate their results through any other technique.
Discussion
CRC is one of the most common types of cancer, which infects millions of people worldwide each year [92]. Although recent advancements in the diagnosis and treatment of the CRC have helped to manage the disease, the prevalence of CRC is still on the rise due to unknown underlying mechanisms [93].
To date, various individual studies have been carried out worldwide to find the state of association between HPV and CRC to further uncover the molecular pathways regulating CRC, but their results are conflicting. In addition, the statistical meta-analysis was also used by the researchers to analyse the previous individual studies for generating a more meaningful association between HPV and CRC, but, due to the shortcomings of statistical meta-analysis, researchers once again failed to establish a more reliable causal relationship between HPV and CRC.
In the present study, we evaluated the previous studies using a reliable, Bradford Hill criteria to find a causal relationship between HPV and CRC. In addition, we also evaluated the methodologies used by the previous studies to address the possibility of false-positive and false-negative results for better outcomes.
In total, 59 original articles were included in the present study. The HPV positivity ratio reported in these studies was varied between 0% [11, 12, 17, 23, 26, 32, 41, 43, 45, 46, 50, 59] and 100% [13, 54]. In most of the case–control studies [12, 17–27, 29–32, 35–57], the positivity ratio for HPV detection was higher in the cancerous samples as compared to the controls, whereas, in some studies [28, 33, 34, 58], HPV positivity ratio was higher in the controls as compared to the CRC samples. Possible reasons for such population-specific inequalities in HPV detection could be non-modifiable factors such as genetic makeup and socially controllable factors such as health-seeking behaviour and differential access to the health facilities. The HPV aetiology in CRC has been discussed population wise as follows:
HPV aetiology in CRC patients of the United States of America (USA)
In the USA, a total of seven studies [11, 21, 22, 26, 33, 51, 64] including four [21, 22, 26, 33] case–control studies have been conducted so far to find out the association between HPV and CRC. These studies used immunohistochemistry and PCR techniques with primers specific for E6, E7 and L1 regions of the viral genome for HPV detection and documented differential HPV detection positivity ratios varying between 0% [21, 22] and 82% [51] in CRC samples, whereas 8% [33] and 38% [33] in normal and adjacent/benign controls, respectively. In the US population, HPV strain 16 was the most frequently reported strain (Table 1).
HPV aetiology in CRC patients of China
In the Chinese population, a total of n = 8 [24, 29, 42, 47–49, 53, 56] case–control studies have carried out up to now reporting an association between HPV and CRC. All of these studies utilised gene chip technology, immunohistochemistry and PCR techniques for the detection of HPV using E6, E7 and L1 region-specific primers and documented varying HPV detection positivity ratios ranging from 21.9% [47] to 73% [42] in CRC samples. On the other side of the coin, they also documented 0% [29, 49] HPV detection positivity in normal and 0% [42]–29.7% [24] in adjacent/benign controls. Hence, their results revealed the higher HPV detection positivity ratios in CRC samples as compared to the normal and adjacent/benign controls. HPV strains 16, 6 and 33 were the most commonly identified strains in the Chinese population (Table 1).
HPV aetiology in CRC patients of India
In India, a single case–control study [27] has been reported so far to find out the association between HPV and CRC. They analysed 93 CRC samples and 30 adjacent/benign controls using L1 gene-specific primers through the PCR technique and identified 6% HPV detection positivity ratio in normal and 36.5% positivity ratio in the CRC samples. The HPV18 was the most prevalent identified strain in the Indian population (Table 1).
HPV aetiology in CRC patients of Iran
In Iran, so far, n = 8 studies [17, 30, 37, 39, 43, 75, 79, 80] have been reported to elaborate the HPV aetiology in CRC. From them, only n = 3 studies [30, 39, 43] were the case–control studies. All these studies employed the PCR technique for HPV detection using primers specific for E6, E7 and L1 region of the viral genome. In this population, HPV detection positivity ratios were reported in varying frequencies ranging from 0% to 35% in CRC samples, whereas 0% [30, 43]–1.25% [39] and 0% [17, 43]–5.7% [30] in normal and adjacent/benign samples, respectively. The HPV strains 16, 18, 55 and 56 were the most commonly identified strains in the Iranian population (Table 1).
Table 1. Summary of the detection of HPV types and positivity rate in normal and CRC samples relative to the different selected articles.
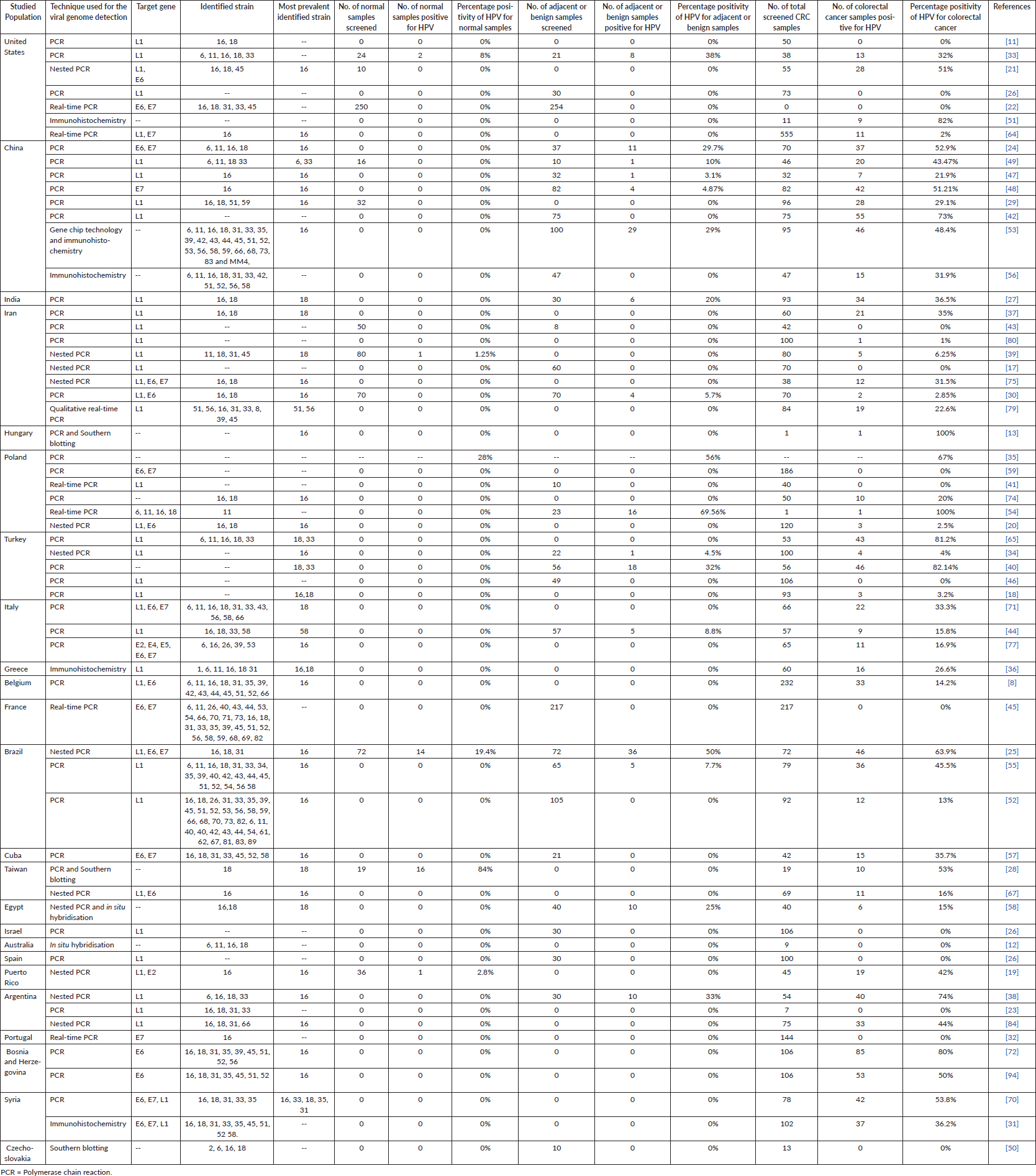
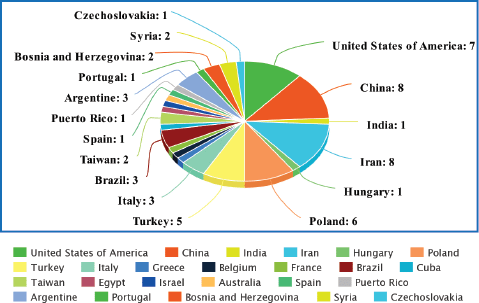
Figure 2. Comparison graph between the numbers (No.) of the studies carried out in each population on HPV in the CRC.
HPV aetiology in CRC patients of Hungary
In Hungary, a single study [13] has been reported to date relating HPV with CRC. They screened only 01 CRC sample using Southern blot hybridisation techniques, and it was positive for HPV 16 (Table 1).
HPV aetiology in CRC patients of Poland
To date, a total of n = 6 studies [20, 35, 41, 54, 59, 74] including n = 2 [41, 54] case–control studies have been carried out in Poland to determine the association between HPV and CRC. HPV detection was done in these studies through the PCR technique using primers specific for E6, E7 and L1 region of the HPV genome. They documented the HPV detection positivity ratios with varying frequencies ranging from 0% [41, 59] to 100% [54] in the CRC samples, whereas 28% [35] in normal samples and from 0% [41] to 69.5% [54] in adjacent/benign samples. HPV 16 was the most commonly identified strain in the Poland population (Table 1).
HPV aetiology in CRC patients of Turkey
A total of five studies [18, 34, 40, 46, 65] have been reported in Turkey until now relating HPV with CRC including three [18, 40, 46] case–control studies. They utilised PCR technique for the detection of HPV using L1 region-specific primers and identified HPV detection positivity ratios with varying frequencies ranging from 0% [46] to 82.14% [40] in CRC samples and 0% [46] to 32% [40] in adjacent/benign. The HPV strains 18 and 33 were the most commonly identified strains in the Turkish population (Table 1).
HPV aetiology in CRC patients of Italy
So far, a total of n = 3 studies [44, 71] including a single case–control study [44] have been reported in Italy to demonstrate the relationship between HPV and CRC. They utilised the PCR technique for the detection of HPV using E6, E7 and L1 region-specific primers and identified HPV detection positivity ratios with varying frequencies ranging from 15.8% [44] to 33.3% [71] in CRC, whereas 8.8% [44] in normal samples. In Italian populations, HPV strains 16 and 18 were the most frequently detected strains (Table 1).
HPV aetiology in CRC patients of Greece
Until now, only a single study [36] has been carried out in Greece to elaborate the association between HPV and CRC. In this study, a total of 60 CRC samples were analysed using the immunohistochemistry technique, and 26.6% HPV positivity ratio was documented (Table 1).
HPV aetiology in CRC patients of Belgium
In Belgium, only a single study [8] has been carried out so far to find the association between HPV with human CRC. In this study, a total of 232 samples were screened using the PCR technique with specific primers targeting the L1 and E6 genes of the HPV genome, and their results revealed a 14.2% HPV detection positivity ratio (Table 1).
HPV aetiology in CRC patients of France
Until now, in France, a single study [45] has been carried out to illustrate the association between HPV and CRC. In total, 217 CRC and adjacent/benign samples were screened using PCR, and HPV was not detected in any of the CRC or control samples (Table 1).
HPV aetiology in CRC patients of Brazil
A couple of case–control studies [25, 52, 55] has been carried out in Brazil so far to analyse the association between HPV and CRC. They utilised PCR with primers specifically targeting the L1, E6 or E7 region of the viral genome and documented HPV detection positivity ratios with varying frequencies ranging from 13% [52] to 63.9% [25] in CRC samples, whereas 19.4% [25] in normal and 0% [52]–50% [25] in adjacent/benign controls. In the Brazilian population, HPV16 was the most commonly identified strain (Table 1).
HPV aetiology in CRC patients of Cuba
Up until now, only a single case–control study [57] relating HPV with CRC has been reported in Cuba. They analysed 42 CRC and 21 adjacent/benign controls using PCR with primers specifically targeting the E6 and E7 regions of the HPV genome and documented 35.7% HPV detection positivity ratio in CRC samples only (Table 1).
HPV aetiology in CRC patients of Taiwan
A couple of studies [28, 67] including a case–control [28] study has been carried out so far in Taiwan to find the association between HPV and CRC. They utilised Southern blotting and PCR for HPV detection and found a varying frequency of HPV detection ranging from 16% [67] to 53% [28] in CRC, whereas 0% in control samples. HPV 16 and 18 were the most commonly identified strains in the Taiwan population (Table 1).
HPV aetiology in CRC patients of Egypt
A single case–control study based on HPV aetiology in CRC has been reported in Egypt so far. They examined 40 CRC as well as adjacent/benign controls using PCR and in situ hybridisation technique and documented 15% and 25% HPV detection positivity ratios in CRC and adjacent/benign controls, respectively (Table 1).
HPV aetiology in CRC patients of Israel
In Israel, there has been a single case–control study [26] carried out so far to determine whether the HPV is associated with CRC or not. They analysed 106 CRC and 30 adjacent/benign controls for HPV detection using PCR and documented a 0% HPV detection positivity ratio (Table 1).
HPV aetiology in CRC patients of Australia
Until now, a single study [12] has been carried out in Australia to demonstrate the relationship between HPV and CRC. They screened only 9 CRC samples using in situ hybridisation technique, and the presence of HPV was not detected in any of the sample (Table 1).
HPV aetiology in CRC patients of Spain
Only a single case–control study [26] has been reported until now in Spain to relate HPV with CRC. In this study, a total of 100 CRC and 30 adjacent/benign controls were analysed using PCR with primers specific for the L1 region of the viral genome, and they observed no HPV marker (Table 1).
HPV aetiology in CRC patients of Puerto Rico
A single case–control study [19] has been carried out in Puerto Rico so far to relate HPV with CRC. In total, 45 CRC and 36 normal samples were analysed in this study using the PCR technique, and they observed 42% and 2.8% HPV detection positivity ratio in CRC and normal controls, respectively (Table 1).
HPV aetiology in CRC patients of Argentine
So far, in total, three studies [38, 58, 84] including a single case–control study [38] have been reported in Argentine relating HPV with CRC. All the studies utilised PCR with primers specifically targeting the L1 region of the viral genome and documented HPV detection positivity ratios with varying frequencies ranging from 0% [23] to 74% [38] and 33% [38] in adjacent/benign samples (Table 1), respectively.
HPV aetiology in CRC patients of Portugal
Only a single study [32] has been carried out in Portugal to date to relate the HPV with human CRC. In total, 144 CRC samples were screened in this study using PCR with primers specific for the E7 region of the viral genome and showed a 0% HPV detection (Table 1).
HPV aetiology in CRC patients of Bosnia and Herzegovina
A total of two studies [72, 94] have been reported in Bosnia and Herzegovina so far relating HPV with the CRC. For the detection of HPV, both the studies utilised PCR with specific primers targeting the E6 region of the viral genome and documented the presence of HPV with varying frequencies ranging from 50% (72) to 80% [94] in CRC samples. The most frequently identified strain in Bosnia and Herzegovina was HPV 16 (Table 1).
HPV aetiology in CRC patients of Syria
So far, in Syria, the association between HPV and CRC has been accessed by only two studies [31, 70]. These studies utilised immunohistochemistry and PCR techniques for HPV detection with primers specific for L1, E6 and E7 regions of the viral genome and documented HPV detection positivity ratios in varying frequencies ranging from 36.2% [31] to 53.8% [70] in CRC samples (Table 1).
HPV aetiology in CRC patients of Czechoslovakia
To date, a single case–control study [50] based on HPV aetiology in CRC has been carried out in Czechoslovakia. A total of 13 cancerous and 10 adjacent/benign samples were analysed in this study using the Southern blot hybridisation technique, and they documented 0% HPV positivity ratio in CRC and adjacent/benign samples (Table 1).
The careful evaluation of the results of identified studies through Bradford Hill criteria showed that all the studies failed to fulfil the major postulates including strength, temporality, consistency, biological gradient, experiment, coherence, specificity and analogy. Hence, we suggested that HPV acts as a coparticipant in the development of CRC rather than having a causal relationship that might combine with the other viruses, such as human immunodeficiency virus and hepatitis C virus and other factors including genetic abnormalities, smoking, alcohol consumption and obesity and may increase a person’s risk of developing CRC by affecting the body’s immune system.
However, limitations and some of the major issues related to methodologies used in the included studies have been discussed as follows.
Possible causes of the false-negative results
Some studies failed to detect HPV presence in any of the cancerous or normal controls that they were investigating. How we can be sure that negative results for HPV detection in the investigating samples are not due to the poor quality of the extracted DNA? Many studies utilised positive control to avoid such situations [17, 30, 39, 43, 80], but few studies [18, 24, 34, 65] did not use positive control, so there is no way to confirm their negative results. The choice of inappropriate primers could lead to false-negative results, for example, primers that target L1 region of the HPV genome may be unreliable for the detection of HPV in tissues of advanced carcinomas, as L1 and E1 regions may be lost during the integration of viral genome into the host genome, whereas the E6/E7 regions remained consistently present, so this is the plausible explanation for the completely negative results of the studies [17, 26, 41, 43, 46].
Possible causes of the false-positive results
Most of the studies that we summarised used PCR technique [8, 13, 18-20, 24, 25, 27-31, 34–40, 42, 44, 47–50, 52, 54, 55, 57, 58, 64–66, 70-72, 74, 75, 77, 79, 80, 84] for the detection of HPV, and none of them utilised second technique to confirm their positive results PCR, except one study [58] which utilised immunohistochemistry. The results of their second technique have deviated from the first one. The expression of the p16, p53 and some other genes could be used as a surrogate biomarker in HPV-infected CRC patients. Along with HPV detection, these surrogate biomarkers were also analysed by some studies [20, 27, 37, 47, 75] to further validate their findings, of which the studies of Laskar et al [27] and Karbasi et al [75] have validated their findings by analysing these surrogate biomarkers, whereas the other studies [20, 37, 47] were failed to validate their findings the surrogate biomarkers. Such deviations in the results of previous studies raise a big question mark on the selection of appropriate technique and their sensitivities.
Comparison of normal, benign and malignant samples
The case–control studies are necessary to establish a causal relationship between the causative agent and the disease. Some of the studies that we summarised analysed only the CRC samples [8, 11, 13, 59, 64, 65, 67, 71, 74, 75, 79, 80, 84] and did not allow us to compare their results with normal or adjacent/benign controls. However, most of the studies also analysed the normal and adjacent/benign tissues along with CRC samples, and the comparison of their results demonstrated that HPV detection positivity ratios in CRC samples were higher in [12, 17, 27, 29–32, 35–57] studies, whereas lower in [28, 33, 34, 58] studies as compared to the normal and adjacent/benign controls. However, none of the studies has reported the association of HPV with specific CRC subtype and histologic grade.
Conclusion
The results of this comprehensive review are controversial. They failed to prove the causal relationship between HPV and CRC, rather suggesting that it is a coparticipant in the pathogenesis of CRC. However, due to various limitations of the methodologies used by the previous studies to detect the presence of HPV in CRC, additional experiments are required to prove the HPV aetiology in CRC.
List of abbreviations
CRC = colorectal cancer
E2 = envelope gene 2
E6 = envelope gene 6
E7 = envelope gene 7
TP53 = tumour suppressor protein 53
MeSH = Medical Subject Headings
References
1. Doosti A, Zamani M, and Dehkordi PG, et al (2011) Association of the p53 codon 72 polymorphism with colorectal cancer in South West of Iran Sci Res Essays 6(15) 3148–3152
2. Giardiello FM, Gurbuz AK, and Bayless TM, et al (1996) Colorectal cancer in ulcerative colitis: survival in patients with and without colorectal cancer symptoms Inflamm Bowel Dis 2(1) 6–10 https://doi.org/10.1097/00054725-199603000-00002 PMID: 23282450
3. Armaghany T, Wilson JD, and Chu Q, et al (2012) Genetic alterations in colorectal cancer Gastrointest Cancer Res 5(1) 19 PMID: 22574233 PMCID: 3348713
4. Siegel RL, Miller KD, and Goding Sauer A, et al (2020) Colorectal cancer statistics, 2020 CA Cancer J Clin 70(3) 145–164 https://doi.org/10.3322/caac.21601 PMID: 32133645
5. Haggar FA and Boushey RP (2009) Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors Clin Colon Rectal Surg 22(4) 191–197 https://doi.org/10.1055/s-0029-1242458 PMCID: 2796096
6. Burnett-Hartman AN, Newcomb PA, and Potter JD (2008) Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus Cancer Epidemiol Biomarkers Prev 17(11) 2970–2979 https://doi.org/10.1158/1055-9965.EPI-08-0571 PMID: 18990738 PMCID: 2676114
7. Burd EM (2003) Human papillomavirus and cervical cancer Clin Microbiol Rev 16(1) 1–17 https://doi.org/10.1128/CMR.16.1.1-17.2003 PMID: 12525422 PMCID: 145302
8. Deschoolmeester V, Van Marck V, and Baay M, et al (2010) Detection of HPV and the role of p16INK4A overexpression as a surrogate marker for the presence of functional HPV oncoprotein E7 in colorectal cancer BMC Cancer 10 117 https://doi.org/10.1186/1471-2407-10-117 PMID: 20346145 PMCID: 2868049
9. Buyru N, Tezol A, and Dalay N (2005) p53 intronic G13964C variant in colon cancer and its association with HPV Anticancer Res 25(4) 2767–2769 PMID: 16080524
10. Dadashi M, Vaezjalali M, and Fallah F, et al (2017) Epidemiology of human papillomavirus (HPV) infection among Iranian women identified with cervical infections: a systematic review and meta-analysis of national data Infect Epidemiol Microbiol 3(2) 68–72
11. Shah KV, Daniel RW, and Simons JW, et al (1992) Investigation of colon cancers for human papillomavirus genomic sequences by polymerase chain reaction J Surg Oncol 51(1) 5–7 https://doi.org/10.1002/jso.2930510104 PMID: 1325576
12. Kulski JK, Demeter T, and Mutavdzic S, et al (1990) Survey of histologic specimens of human cancer for human papillomavirus types 6/11/16/18 by filter in situ hybridization Am J Clin Pathol 94(5) 566–570 https://doi.org/10.1093/ajcp/94.5.566 PMID: 2173398
13. Bognár G, István G, and Bereczky B, et al (2008) Detection of human papillomavirus type 16 in squamous cell carcinoma of the colon and its lymph node metastases with PCR and southern blot hybridization Pathol Oncol Res 14(1) 93–96 https://doi.org/10.1007/s12253-008-9011-6 PMID: 18350376
14. Damin DC, Ziegelmann PK, and Damin AP (2013) Human papillomavirus infection and colorectal cancer risk: a meta-analysis Colorectal Dis 15(8) 12257 https://doi.org/10.1111/codi.12257
15. Baandrup L, Thomsen LT, and Olesen TB, et al (2014) The prevalence of human papillomavirus in colorectal adenomas and adenocarcinomas: a systematic review and meta-analysis Eur J Cancer 50(8) 1446–1461 https://doi.org/10.1016/j.ejca.2014.01.019 PMID: 24560489
16. Fedak KM, Bernal A, and Capshaw ZA, et al (2015) Applying the bradford hill criteria in the 21st century: how data integration has changed causal inference in molecular epidemiology Emerg Themes Epidemiol 12 14 https://doi.org/10.1186/s12982-015-0037-4 PMID: 26425136 PMCID: 4589117
17. Aghakhani A, Hamkar R, and Ramezani A, et al (2014) Lack of human papillomavirus DNA in colon adenocarcinama and adenoma J Cancer Res Ther 10(3) 531–534 PMID: 25313733
18. Balta AZ, Berber U, and Tanoglu A, et al (2014) The potential role of human papillomavirus in colorectal carcinoma Asian Pac J Cancer Prev 15(10):4375 https://doi.org/10.7314/APJCP.2014.15.10.4375 PMID: 24935400
19. Bernabe-Dones RD, Gonzalez-Pons M, and Villar-Prados A, et al (2016) High prevalence of human papillomavirus in colorectal cancer in hispanics: a case-control study Gastroenterol Res Pract 2016 7896716 https://doi.org/10.1155/2016/7896716 PMID: 26904111 PMCID: 4745930
20. Biesaga B, Janecka-Widła A, and Kołodziej-Rzepa M, et al (2019) The prevalence of HPV infection in rectal cancer—report from South—Central Poland (Cracow region) Pathol Res Pract 215(9) 28 https://doi.org/10.1016/j.prp.2019.152513
21. Bodaghi S, Yamanegi K, and Xiao S-Y, et al (2005) Colorectal papillomavirus infection in patients with colorectal cancer Clin Cancer Res 11(8) 2862–2867 https://doi.org/10.1158/1078-0432.CCR-04-1680 PMID: 15837733 PMCID: 1479314
22. Burnett-Hartman AN, Newcomb PA, and Mandelson MT, et al (2011) No evidence for human papillomavirus in the etiology of colorectal polyps Cancer Epidemiol Biomark Prev 20(10) 2288–2297 https://doi.org/10.1158/1055-9965.EPI-11-0450
23. Cavatorta AL, Fumero G, and Chouhy D, et al (2004) Differential expression of the human homologue of drosophila discs large oncosuppressor in histologic samples from human papillomavirus-associated lesions as a marker for progression to malignancy Int J Cancer 111(3) 373–380 https://doi.org/10.1002/ijc.20275 PMID: 15221964
24. Cheng JY, Sheu LF, and Meng CL, et al (1995) Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction Gut 37(1) 87–90 https://doi.org/10.1136/gut.37.1.87 PMID: 7672688 PMCID: 1382774
25. Damin DC, Caetano MB, and Rosito MA, et al (2007) Evidence for an association of human papillomavirus infection and colorectal cancer Eur J Surg Oncol 33(5) 569–574 https://doi.org/10.1016/j.ejso.2007.01.014 PMID: 17321098
26. Gornick MC, Castellsague X, and Sanchez G, et al (2010) Human papillomavirus is not associated with colorectal cancer in a large international study Cancer Causes Control 21(5) 737–743 https://doi.org/10.1007/s10552-010-9502-0 PMID: 20087645 PMCID: 4269349
27. Laskar RS, Talukdar FR, and Choudhury JH, et al (2015) Association of HPV with genetic and epigenetic alterations in colorectal adenocarcinoma from Indian population Tumour Biol 36(6) 4661–4670 https://doi.org/10.1007/s13277-015-3114-y PMID: 25647260
28. Lee YM, Leu SY, and Chiang H, et al (2001) Human papillomavirus type 18 in colorectal cancer J Microbiol Immunol Infect 34(2) 87–91 PMID: 11456365
29. Liu F, Mou X, and Zhao N, et al (2011) Prevalence of human papillomavirus in Chinese patients with colorectal cancer Colorectal Dis 13(8) 865–871 https://doi.org/10.1111/j.1463-1318.2010.02335.x
30. Mahmoudvand S, Safaei A, and Erfani N, et al (2015) Presence of human papillomavirus DNA in colorectal cancer tissues in Shiraz, Southwest Iran Asian Pac J Cancer Prev 16(17) 7883–7887 https://doi.org/10.7314/APJCP.2015.16.17.7883 PMID: 26625815
31. Malki MI, Gupta I, and Fernandes Q, et al (2020) Co-presence of Epstein–Barr virus and high-risk human papillomaviruses in Syrian colorectal cancer samples Hum Vaccin Immunother 1–5 https://doi.org/10.1080/21645515.2020.1726680
32. Martins SF, Mariano V, and Rodrigues M, et al (2020) Human papillomavirus (HPV) 16 infection is not detected in rectal carcinoma Infect Agent Cancer 15(1) 17 https://doi.org/10.1186/s13027-020-00281-z PMID: 32165915 PMCID: 7059378
33. McGregor B, Byrne P, and Kirgan D, et al (1993) Confirmation of the association of human papillomavirus with human colon cancer Am J Surg 166(6) 738–740 https://doi.org/10.1016/S0002-9610(05)80690-7 PMID: 8273860
34. Militello V, Trevisan M, and Squarzon L, et al (2009) Investigation on the presence of polyomavirus, herpesvirus, and papillomavirus sequences in colorectal neoplasms and their association with cancer Int J Cancer 124(10) 2501–2503 https://doi.org/10.1002/ijc.24224 PMID: 19170205
35. Młynarczyk B, Malejczyk M, and Muszyński J, et al (2009) The occurrence of human papillomavirus--HPV in the biopsies from colon polyps and cancer Med Dosw Mikrobiol 61(2) 191–196
36. Moreas H, Tsiambas E, and Lazaris AC, et al (2014) Impact of HPV detection in colorectal adenocarcinoma: HPV protein and chromogenic in situ hybridization analysis based on tissue microarrays J BUON 19(1) 91–96 PMID: 24659648
37. Motlagh A, Azadeh P, and Hashemi M, et al (2012) Human papillomavirus infection, p53 overexpression and histopathologic characteristics in colorectal cancer Govaresh 12(2) 126–133
38. Pérez LO, Abba MC, and Laguens RM, et al (2005) Analysis of adenocarcinoma of the colon and rectum: detection of human papillomavirus (HPV) DNA by polymerase chain reaction Colorectal Dis 7(5) 492–495 https://doi.org/10.1111/j.1463-1318.2005.00774.x PMID: 16108887
39. Ranjbar R, Saberfar E, and Shamsaie A, et al (2014) The aetiological role of human papillomavirus in colorectal carcinoma: an Iranian population-based case control study Asian Pac J Cancer Prev 15(4) 1521–1525 https://doi.org/10.7314/APJCP.2014.15.4.1521 PMID: 24641361
40. Salepci T, Yazici H, and Dane F, et al (2009) Detection of human papillomavirus DNA by polymerase chain reaction and southern blot hybridization in colorectal cancer patients J BUON 14(3) 495–499 PMID: 19810144
41. Snietura M, Waniczek D, and Nowakowska-Zajdel E, et al (2012) Does human papilloma virus participate in colorectal carcinogenesis? J Biol Regul Homeost Agents 26(4) 757–762 PMID: 23241125
42. Sun Z-Q, Wang H-J, and Zhao Z-L, et al (2013) Significance of HPV infection and genic mutation of APC and K-ras in patients with rectal cancer Asian Pac J Cancer Prev 14(1) 121–126 https://doi.org/10.7314/APJCP.2013.14.1.121 PMID: 23534709
43. Taherian H, Tafvizi F, and Fard ZT, et al (2014) Lack of association between human papillomavirus infection and colorectal cancer Prz Gastroenterol 9(5) 280–284 PMID: 25396002 PMCID: 4223116
44. Tanzi E, Bianchi S, and Frati ER, et al (2015) Human papillomavirus detection in paraffin-embedded colorectal cancer tissues J Gen Virol 96(Pt 1) 206–209 https://doi.org/10.1099/vir.0.070672-0
45. Vuitton L, Jaillet C, and Jacquin E, et al. (2017) Human papillomaviruses in colorectal cancers: a case-control study in western patients Dig Liver Dis 49(4) 446–450 https://doi.org/10.1016/j.dld.2016.11.003
46. Yavuzer D, Karadayi N, and Salepci T, et al (2011) Investigation of human papillomavirus DNA in colorectal carcinomas and adenomas Med Oncol (Northwood, London, England) 28(1) 127–132 https://doi.org/10.1007/s12032-010-9416-4
47. Yu H-G, Shun L-B, and Luo H-S, et al (2002) Deletion of the FHIT gene in human colorectal cancer is independent of high-risk HPV infection Int J Colorectal Dis 17(6) 396–401 https://doi.org/10.1007/s00384-002-0404-9 PMID: 12355215
48. Zhang J, Ding Y, and Zhou Z, et al (2005) Expression of human papillomavirus 16 E7 DNA in patients with colorectal adenocarcinoma Sheng wu yi xue gong cheng xue za zhi 22(5) 1024–1044 PMID: 16294745
49. Zhu Q, Cao J, and Li S (1999) Detection of human papillomavirus gene in biopsies from colon carcinoma by PCR Zhonghua shi yan he lin chuang bing du xue za zhi 13(4) 352–354
50. Boguszaková L, Hirsch I, and Brichácek B, et al (1988) Absence of cytomegalovirus, Epstein-Barr virus, and papillomavirus DNA from adenoma and adenocarcinoma of the colon Acta Virol 32(4) 303–308 PMID: 2903634
51. Cimino-Mathews A, Sharma R, and Illei PB (2012) Detection of human papillomavirus in small cell carcinomas of the anus and rectum Am J Surg Pathol 36(7) 1087–1092 https://doi.org/10.1097/PAS.0b013e3182549b6d PMID: 22531171
52. Dalla Libera LS, de Siqueira T, and Santos IL, et al (2020) Detection of human papillomavirus and the role of p16INK4a in colorectal carcinomas PLoS One 15(6) e0235065 https://doi.org/10.1371/journal.pone.0235065 PMID: 32584870 PMCID: 7316293
53. Li YX, Zhang L, and Simayi D, et al (2015) Human papillomavirus infection correlates with inflammatory Stat3 signaling activity and IL-17 level in patients with colorectal cancer PLoS One 10(2) e0118391 https://doi.org/10.1371/journal.pone.0118391 PMID: 25706309 PMCID: 4338045
54. Mlynarczyk-Bonikowska B, Muszyński J, and Szymanek-Mjchrzak K, et al (2017) The prevalence of human papillomaviruses in patients with colon polyps Med Dosw Mikrobiol 69(1) 49–54 PMID: 30351624
55. Picanço-Junior OM, Oliveira AL, and Freire LT, et al (2014) Association between human papillomavirus and colorectal adenocarcinoma and its influence on tumor staging and degree of cell differentiation Arq Bras Cir Dig 27(3) 172–176 https://doi.org/10.1590/S0102-67202014000300003 PMID: 25184765 PMCID: 4676375
56. Qiu Q, Li Y, and Fan Z, et al (2020) Gene expression analysis of human papillomavirus-associated colorectal carcinoma Biomed Res Int 2020 5201587 https://doi.org/10.1155/2020/5201587 PMID: 32258125 PMCID: 7103040
57. Soto Y, Limia CM, and González L, et al (2016) Molecular evidence of high-risk human papillomavirus infection in colorectal tumours from Cuban patients Mem Inst Oswaldo Cruz 111(12) 731–736 https://doi.org/10.1590/0074-02760160217 PMID: 27812599 PMCID: 5146735
58. El-Seidi EA, Sorour AE, and Gamil M, et al (2014) Human papillomavirus in patients with colorectal cancer Egypt J Med Microbiol 23(1)
59. Karpinski P, Myszka A, and Ramsey D, et al (2011) Detection of viral DNA sequences in sporadic colorectal cancers in relation to CpG island methylation and methylator phenotype Tumour Biol 32(4) 653–659 https://doi.org/10.1007/s13277-011-0165-6 PMID: 21625944 PMCID: 3131518
60. Arnold M, Sierra MS, and Laversanne M, et al (2017) Global patterns and trends in colorectal cancer incidence and mortality Gut 66(4) 683–691 https://doi.org/10.1136/gutjnl-2015-310912
61. Banno K, Iida M, and Yanokura M, et al (2014) MicroRNA in cervical cancer: oncomiRs and tumor suppressor miRs in diagnosis and treatment Sci World J 2014 178075 https://doi.org/10.1155/2014/178075
62. Boland CR, Bigler J, and Newcomb PA, et al (2004) Evidence for an association between JC virus and colorectal neoplasia Cancer Epidemiol Biomarkers Prev 13(12) 2285–2286 PMID: 15598796
63. Buitrago-Pérez Á, Garaulet G, and Vázquez-Carballo A, et al (2009) Molecular signature of HPV-induced carcinogenesis: pRb, p53 and gene expression profiling Curr Genomics 10(1) 26–34 https://doi.org/10.2174/138920209787581235 PMID: 19721808 PMCID: 2699838
64. Burnett-Hartman AN, Feng Q, and Popov V, et al (2013) Human papillomavirus DNA is rarely detected in colorectal carcinomas and not associated with microsatellite instability: the Seattle colon cancer family registry Cancer Epidemiol Biomarkers Prev 22(2) 317–319 https://doi.org/10.1158/1055-9965.EPI-12-1170 PMCID: 3565050
65. Buyru N, Tezol A, and Dalay N (2006) Coexistence of K-ras mutations and HPV infection in colon cancer BMC Cancer 6 115 https://doi.org/10.1186/1471-2407-6-115 PMID: 16672071 PMCID: 1481576
66. Chan DS, Lau R, and Aune D, et al (2011) Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies PLoS One 6(6) e20456 https://doi.org/10.1371/journal.pone.0020456 PMID: 21674008 PMCID: 3108955
67. Chen T-H, Huang C-C, and Yeh K-T, et al (2012) Human papilloma virus 16 E6 oncoprotein associated with p53 inactivation in colorectal cancer World J Gastroenterol 18(30) 4051–4058 https://doi.org/10.3748/wjg.v18.i30.4051 PMID: 22912557 PMCID: 3420003
68. Coelho TR, Almeida L, and Lazo PA (2010) JC virus in the pathogenesis of colorectal cancer, an etiological agent or another component in a multistep process? Virol J 7(1) 42 https://doi.org/10.1186/1743-422X-7-42 PMID: 20167111 PMCID: 2830963
69. Fedrizzi EN (2011) Epidemiologia da infecção genital pelo HPV Rev Bras Pat Trato Gen Inf 1(1) 3–8
70. Ghabreau L, Segal ED, and Yasmeen A, et al (2012) High-risk human papillomavirus infections in colorectal cancer in the Syrian population and their association with Fascin, Id-1 and P-cadherin expressions: a tissue microarray study Clin Cancer Investig J 1(1) 26 https://doi.org/10.4103/2278-0513.95016
71. Giuliani L, Ronci C, and Bonifacio D, et al (2008) Detection of oncogenic DNA viruses in colorectal cancer Anticancer Res 28(2B) 1405–1410 PMID: 18505087
72. Gupta I, Al Farsi H, and Jabeen A, et al (2020) High-risk human papillomaviruses and Epstein–Barr virus in colorectal cancer and their association with clinicopathological status Pathogens 9(6) 452 https://doi.org/10.3390/pathogens9060452 PMCID: 7350373
73. Höcker M and Hohenberger P (2003) Helicobacter pylori virulence factors—one part of a big picture Lancet 362(9391) 1231–1233 https://doi.org/10.1016/S0140-6736(03)14547-3 PMID: 14568748
74. Jarzyński A, Zając P, and Żebrowski R, et al (2017) Occurrence of BK virus and human papilloma virus in colorectal cancer Ann Agric Environ Med 24(3) 440–445 https://doi.org/10.26444/aaem/74648
75. Karbasi A, Borhani N, and Daliri K, et al (2015) Downregulation of external death receptor genes FAS and DR5 in colorectal cancer samples positive for human papillomavirus infection Pathol Res Pract 211(6) 444–448 https://doi.org/10.1016/j.prp.2015.02.001 PMID: 25795228
76. Liang PS, Chen T-Y, and Giovannucci E (2009) Cigarette smoking and colorectal cancer incidence and mortality: systematic review and meta-analysis Int J Cancer 124(10) 2406–2415 https://doi.org/10.1002/ijc.24191 PMID: 19142968
77. Lorenzon L, Mazzetta F, and Pilozzi E, et al (2015) Human papillomavirus does not have a causal role in colorectal carcinogenesis World J Gastroenterol 21(1) 342–350 https://doi.org/10.3748/wjg.v21.i1.342 PMID: 25574110 PMCID: 4284354
78. Lynch HT and De la Chapelle A (2003) Hereditary colorectal cancer N Engl J Med 348(10) 919–932 https://doi.org/10.1056/NEJMra012242 PMID: 12621137
79. Malekpour Afshar R, Deldar Z, and Mollaei HR, et al (2018) Evaluation of HPV DNA positivity in colorectal cancer patients in Kerman, Southeast Iran Asian Pac J Cancer Prev 19(1) 193–198 PMID: 29373913 PMCID: 5844617
80. Meshkat M, Tayyebi Meibodi N, and Sepahi S, et al (2014) The frequency of human papillomaviruses in colorectal cancer samples in Mashhad, northeastern Iran Turk J Med Sci 44(3) 501–503 https://doi.org/10.3906/sag-1303-81
81. Muñoz N (2000) Human papillomavirus and cancer: the epidemiological evidence J Clin Virol 19(1–2) 1–5 https://doi.org/10.1016/S1386-6532(00)00125-6 PMID: 11091143
82. Murphy G, Pfeiffer R, and Camargo MC, et al (2009) Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location Gastroenterology 137(3) 824–833 https://doi.org/10.1053/j.gastro.2009.05.001 PMID: 19445939 PMCID: 3513767
83. Panic N, Leoncini E, and de Belvis G, et al (2013) Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses PLoS One 8(12) e83138 https://doi.org/10.1371/journal.pone.0083138
84. Pérez LO, Barbisan G, and Ottino A, et al (2010) Human papillomavirus DNA and oncogene alterations in colorectal tumors Pathol Oncol Res 16(3) 461–468 https://doi.org/10.1007/s12253-010-9246-x PMID: 20232185
85. Souto R, Falhari JPB, and Cruz A (2005) O papilomavírus humano: um fator relacionado com a formação de neoplasias Rev Bras Cancerol 51(2) 155–160
86. Souza DL, Jerez-Roig J, and Cabral FJ, et al (2014) Colorectal cancer mortality in Brazil: predictions until the year 2025 and cancer control implications Dis Colon Rectum 57(9) 1082–1089 https://doi.org/10.1097/DCR.0000000000000186 PMID: 25101604
87. Team RC (2013) R: a language and environment for statistical computing
88. Win AK, Walters RJ, and Buchanan DD, et al (2012) Cancer risks for relatives of patients with serrated polyposis Am J Gastroenterol 107(5) 770–778 https://doi.org/10.1038/ajg.2012.52 PMID: 22525305 PMCID: 3488375
89. Yugawa T and Kiyono T (2009) Molecular mechanisms of cervical carcinogenesis by high‐risk human papillomaviruses: novel functions of E6 and E7 oncoproteins Rev Med Virol 19(2) 97–113 https://doi.org/10.1002/rmv.605 PMID: 19156753
90. Zhang Y, Fan S, and Meng Q, et al (2005) BRCA1 interaction with human papillomavirus oncoproteins J Biol Chem 280(39) 33165–33177 https://doi.org/10.1074/jbc.M505124200 PMID: 15983032
91. Zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application Nat Rev Cancer 2(5) 342–350 https://doi.org/10.1038/nrc798 PMID: 12044010
92. Mármol I, Sánchez-de-Diego C, and Pradilla Dieste A, et al (2017) Colorectal carcinoma: a general overview and future perspectives in colorectal cancer Int J Mol Sci 18(1) 197 https://doi.org/10.3390/ijms18010197 PMCID: 5297828
93. Pretzsch E, Bösch F, and Neumann J, et al (2019) Mechanisms of metastasis in colorectal cancer and metastatic organotropism: hematogenous versus peritoneal spread J Oncol 2019 7407190 https://doi.org/10.1155/2019/7407190 PMID: 31641356 PMCID: 6770301
94. Gupta I, Al Farsi H, and Jabeen A, et al (2020) High-risk human papillomaviruses and epstein–barr virus in colorectal cancer and their association with clinicopathological status Pathogens 9(6) 452 https://doi.org/10.3390/pathogens9060452 PMCID: 7350373