Circulating and tissue biomarkers in early-stage non-small cell lung cancer
Caterina Fumagalli1, Fabrizio Bianchi2, Paola Rafaniello Raviele1, Davide Vacirca1, Giovanni Bertalot3, Cristiano Rampinelli4, Matteo Lazzeroni5, Bernardo Bonanni5, Giulia Veronesi6, Nicola Fusco7, Massimo Barberis1 and Elena Guerini-Rocco1
1Division of Pathology, European Institute of Oncology, Via Giuseppe Ripamonti 435, 20141, Milan, Italy
2Institute for Stem-cell Biology, Regenerative Medicine and Innovative Therapies (ISBReMIT), IRCCS Casa Sollievo della Sofferenza, Viale Cappuccini 1, 71013, San Giovanni Rotondo, Foggia, Italy.
3Molecular Medicine Programme IEO, European Institute of Oncology, Via Giuseppe Ripamonti 435, 20141, Milan, Italy
4Department of Radiology, European Institute of Oncology, Via Giuseppe Ripamonti 435, 20141, Milan, Italy
5Division of Cancer Prevention and Genetics, European Institute of Oncology, Via Giuseppe Ripamonti 435, 20141, Milan, Italy
6Division of Thoracic Surgery, Humanitas Research Hospital, Via Manzoni 56, 20089, Rozzano Milan, Italy
7Division of Pathology, Fondazione IRCCS Ca’ Granda – Ospedale Maggiore Policlinico, University of Milan, Via Francesco Sforza 35, 20122, Milan, Italy
Correspondence to: Elena Guerini-Rocco and Massimo Barberis. E-mail: Elena.GueriniRocco@ieo.it and Massimo.Barberis@ieo.it
Abstract
Objective: We sought to characterise circulating and tissue tumour biomarkers of patients who developed early-stage non-small cell lung cancer (NSCLC) during long-term follow-up of a chemoprevention trial (NCT00321893).
Materials and Methods: Blood and sputum samples were collected from 202 high-risk asymptomatic individuals with CT-detected stable lung nodules. Real-time PCR was performed on plasma to quantify free circulating DNA. Baseline serum was investigated with a previously validated test based on 13 circulating miRNAs (miR-Test). Promoter methylation status of p16, RASSF1a and RARβ2 and telomerase activity were assessed in sputum samples. DNA was extracted from each tumour developed during follow-up and subjected to a mutation survey using the LungCarta panel on the Sequenom MassARRAY platform.
Results: During follow-up (9 years) six individuals underwent surgery for stage I NSCLC with a median time of disease onset of 20.5 months. MiR-Test scores were positive (range: 0.14–7.24) in four out of six baseline pre-disease onset sera. No association was identified between free circulating DNA or sputum biomarkers and disease onset. All tumours harboured at least one somatic mutation in well-known cancer genes, including KRAS (n = 4), BRAF (n = 1), and TP53 (n = 3).
Conclusion: Circulating miRNA tests may represent valuable tools to detect clinically-silent tumours. Early-stage lung adenocarcinomas harbour recurrent genetic events similar to those described in advanced-stage NSCLCs.
Keywords: non-small cell lung cancer; early detection; circulating biomarker; somatic mutation
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (
Published: 31/01/2017; Received: 27/07/2016
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related deaths worldwide [1]. Primary prevention based on tobacco control programmes still remains the most effective approach to tackle this highly lethal disease. However, early detection of lung cancer represents a fundamental strategy to reduce disease-associated mortality and allowing for the application of potentially curative treatments (i.e. surgical resection) [2]. Unfortunately, only less than 20% of patients are currently diagnosed with a locally confined disease [3].
Over the past decade, low-dose computed tomography (LDCT) has been shown to be an effective screening tool for early detection of tumours in high-risk populations, reducing lung cancer mortality [4, 5]. However, the majority of the LDCT-detected lung nodules are benign lesions [5, 6], entailing further expensive and invasive procedures. In this scenario, circulating biomarkers might represent the ultimate complementary tools to improve the cost-effectiveness of screening protocols in high-risk populations, decreasing the number of false positive cases, and allowing for non-invasive early diagnosis [7]. Circulating-free DNA (cfDNA) has been extensively investigated as a potential diagnostic, predictive, and prognostic non-invasive biomarker in different tumour types [8–10]. Quantitative and qualitative alterations of cfDNA, including both genetic and epigenetic aberrations, have been described in blood and sputum samples from patients with NSCLCs even at early stages of tumour onset [11–16]. Moreover, the expression profiles of circulating short non-coding microRNA molecules (miRNAs) have been shown to represent a compelling non-invasive biomarker for the diagnosis of cancer [17, 18]. Recently, different studies have reported on the accuracy of different serum and plasma circulating miRNA signatures for early detection of lung tumour [19–23]. In particular, Montani et al identified a serum miRNA signature (miR-Test) based on 13 circulating miRNAs which have been validated in previous lung cancer screening studies demonstrating a high sensitivity and specificity (77.8% and 74.8%, respectively) for early diagnosis of lung cancer [22]. In addition, chemoprevention strategies have been combined with smoking cessation and screening programmes using agents, such as corticosteroids or aspirins, with heterogeneous and still non-conclusive results about their effect of lung cancer risk-reduction [24–26].
In this study, we sought to explore the landscape of circulating and tumour tissue molecular biomarkers of patients who developed early-stage NSCLC during long-term follow-up of randomised, double-blind, phase IIb chemoprevention trial nested in a computed tomography (CT)-scan lung cancer screening programme (NCT00321893) [24].
Materials and methods
Population and samples
Study design and eligibility criteria of the randomised, double-blind, phase IIb chemoprevention trial (NCT00321893) have been extensively described before [27]. Briefly, 202 high-risk asymptomatic subjects, current or former smokers, with CT-detected lung nodules that were persistent for at least one year were randomised to receive budesonide (800 µg) or placebo twice daily for 12 months. Nodule types were classified as non-solid (n = 23), partially solid (n = 41), solid (n = 184), and sub-solid (n = 64). Sputum and blood samples were collected from each individual at baseline and after 6 and 12 months of treatment. Both plasma and serum were obtained from blood samples. Informed consent was obtained as specified in the trial protocols and previously reported [24, 27]. Dropout rate was 2%. Clinical short-term and long-term effects of one year of inhaled budesonide on screening-detected lung nodules have already been reported at a follow-up of one and five years respectively [24, 28]. During follow-up (nine years in total), six patients developed NSCLC.
Circulating biomarker analyses
Promoter methylation status of p16INK4A, RASSF1a, and RARβ2 genes was analysed in sputum samples with quantitative methylation specific PCR (QMSP) as previously described [16, 29, 30]. Telomerase activity assay was performed on sputum using commercial available kit (TeloTAGGG Telomerase PCR ELISAPLUS, Roche) following manufacturer’s protocol. Sputum analyses were performed at baseline and after 12 months from enrolment. Using a real-time quantitative PCR, cfDNA was quantified on plasma samples collected at baseline after 6 and later 12 months from enrolment according to Sozzi et al [31]. The previously validated miR-Test was retrospectively assessed on baseline serum of the six patients who developed NSCLC and fifty cancer-free individuals enrolled in the same trial. Serum miRNA purification and expression profiling were performed according to Montani et al [22]. Briefly, total RNA purification, including miRNAs, was based on lysis with guanidinium thiocyanate-phenol-chloroform extraction (TriZol-LS, Applied Biosystem) and Spin Column-based total RNA purification (MiRneasy Mini Kit, Qiagen). MiRNA qRT-PCR was carried out on a ViiA™ 7 instruments (ThermoFisher) using the manufacturer’s recommended cycling conditions. MiRNA qRT-PCR data were automatically analysed using a custom R script that provides miR-Test risk scores automatically. Patients were classified as ‘positive’ or ‘negative’ for the miR-Test based on a risk score ≥ 0 or < 0 respectively [22].
Tumours pathologic assessment and mutation analysis
Haematoxylin and eosin stained sections of each case were reviewed by two pathologists; tumours were staged and subtyped according to WHO Classification of Tumours of the Lung [32]. DNA was extracted from representative 5-µm-thick sections cut from formalin-fixed and paraffin-embedded blocks of each tumour sample to ensure tumour cell content is above 20% as previously described [33]. Genomic DNAs were subjected to a mutation survey using the LungCarta panel (Sequenom), including evaluation of 214 somatic mutations in 26 oncogenes and tumour suppressor genes, and analysed on a matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometer (Sequenom) following manufacturer’s protocol. Data were evaluated using MassARRAY TYPER ANALYSER software 4.0, with a limit of detection of 5%.
Results
No tumour occurred during the 12 months of trial treatment. Only six patients, three in the budesonide arms and three in the placebo arms, were diagnosed with NSCLC during the following 9 years of follow-up with a median time from enrolment to disease onset of 663.5 days. One of the six patients underwent a local pulmonary recurrence 3 years after the first diagnosis. After a median follow-up of 2104 days from surgery, each patient was alive and disease-free. Patient’s characteristics are reported in Table 1. All tumours were classified as stage I lung adenocarcinomas (ADC) including one case with multifocal minimally-invasive adenocarcinoma (MIA). All lesions displayed a prevalent lepidic and/or acinar pattern of growth (Figure 1). The clinic-pathologic characteristics of tumours and the radiologic features of nodules at baseline and at the time of tumour onset are summarised in Table 1.
No significant differences were observed in plasma cfDNA or sputum biomarkers between patients who developed NSCLC and the whole trial cohort (Table 2). The epigenetic analysis revealed p16INK4A or RASSF1a methylation in 16 samples and none from patients who developed lung cancer.
Interestingly, telomerase activity was negative in all but two sputum samples collected at 12 months after the beginning of the treatment. Both samples were obtained from two of the patients who subsequently developed lung cancer (patients #1 and #3) (Table 2), and the presence of telomerase activity was also confirmed in their corresponding frozen tumour tissues.
The miR-Test revealed a positive score in four out of six baseline pre-disease onset sera (sensitivity 67%) of patients that subsequently were diagnosed with NSCLC (median: range: 0.14– 4.81). Remarkably, the two miR-Test negative patients showed a positive telomerase activity in their sputum samples. Among the 50 cancer-free individuals, 36 had a negative miR Test score (specificity 72%), with a median risk score of -6.2 (range: -21.5 – 28.9) (Table 2).
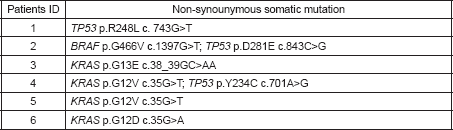
All tumour samples harboured at least one driver somatic mutation in well-known cancer-related genes, including KRAS (n = 4), BRAF (n = 1), and TP53 (n = 3) (Table 3). Interestingly, non-synonymous somatic missense mutations of KRAS were detected in four out of the six (66.67%) early-stage adenocarcinoma analysed with a frequency even higher than that described in early and advanced-stage adenocarcinoma of high-risk individuals (32% and 35% respectively) [34–36].
Table 1. Clinico-pathologic features of baseline nodules and tumors of the six patients who developed lung adenocarcinoma.
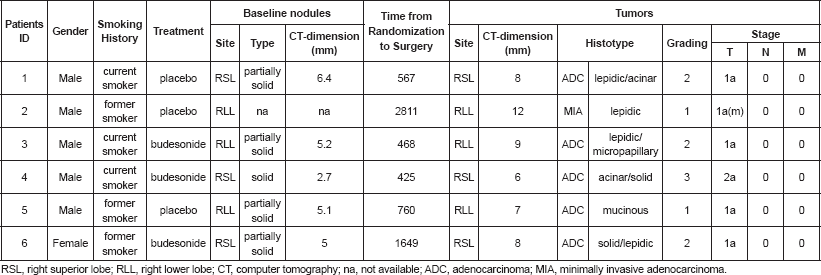
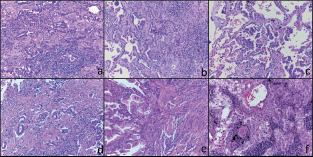
Figure 1. Histological features of the six tumours included in this study. Representative micrographs of the six tumours showing adenocarcinoma with a prevalent lepidic and/or acinar growth pattern (a-f) with focal formation of micropapillae in case 3 (c) and solid nests in case 6 (f). Haematoxylin and eosin-stained slides; original magnification 200x.
Table 2. Circulating biomarkers of the six patients who developed lung adenocarcinoma and median values of the lung cancer-free group.
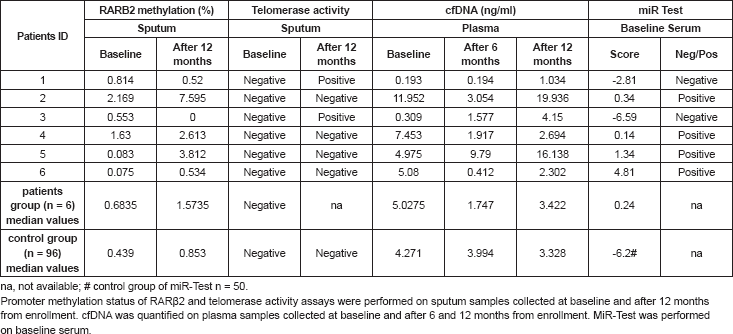
Table 3. Non-synonymous somatic mutations identified in the six lung adenocarcinoma.
Discussion
The continuous monitoring of high-risk individuals enrolled in lung cancer-screening programmes represents a great opportunity not only in detecting early-stage, potentially curable disease but also in validating and/or identifying new diagnostic molecular biomarkers of lung cancer. Here, we performed a multi-level characterisation of circulating and tumour tissue biomarkers of a unique group of patients who developed early-stage NSCLCs during long-term follow-up of a chemoprevention trial (NCT00321893) [24] nested in CT-scan screening programme [37].
Primary and secondary prevention strategies still remain the pivotal steps to reduce the high-rate of NSCLC-associated mortality. Over the past decade, many efforts have been made to identify effective programmes of prevention for high-risk individuals, including chemoprevention and imaging screening studies. In this study, among 202 high-risk asymptomatic subjects with CT-detected lung nodules enrolled in one-year budesonide-based chemoprevention trial, only six patients developed NSCLC during long-term follow-up with a rate slightly lower than that described in the entire nested screening programme population (3% versus 5.7%) [38]. It should be noted that these rates refer specifically to the follow-up timeframe of this study. As previously reported [24, 28], no differences were identified in the distribution of lung cancer between treatment- and placebo-arm. All patients were diagnosed with early-stage NSCLCs and were eligible for surgical resection confirming the efficacy of such screening programmes in the early detection of potentially curable diseases.
The median concentration of plasma cfDNA and sputum RARβ2 gene methylation did not show any statistically significant difference in NSCLC patients compared to the cancer-free group. Moreover, no gene promoter methylation of p16INK4A and RASSF1a was detected in any sputum sample from lung-cancer patients. Furthermore, four tumours harboured KRAS somatic mutations that have been previously shown to have a tendency toward mutual exclusivity with RASSF1a promoter methylation in colorectal and non-small cell lung cancers [39, 40]. These results suggest that plasma cfDNA and sputum gene methylation quantifications, albeit specific [16], are likely to represent low-sensitive diagnostic biomarkers, especially in NSCLC-screening settings.
The sputum telomerase activity assay was able to capture two out six clinically-silent lung tumours. In previous studies, tissue telomerase activity has been correlated with poor-prognostic early and advanced-stage NSCLCs [15, 41]. The identification of telomerase activity in pre-disease onset sputum indicates that it might represent also a potentially useful diagnostic biomarker in early-stage lung cancers. Interestingly, Ilie et al described a sensitive circulating tumour cell (CTC) detection approach to identify patients 'at risk' of developing lung cancer before any clinically detectable CT scan nodules [42]. Additional multicentric studies are warranted to define the specificity and sensitivity of this approach in high-risk asymptomatic subjects with CT-detected lung nodules and to characterise the potential role of these circulating biomarkers as reliable screening tools for early detection of lung cancer in clinical practice.
Since the design of this trial protocol [27] new circulating biomarkers for early-detection of NSCLC have been investigated. In particular, the miR-Test has been recently described as a promising tool to detect clinically-silent tumours [21]. Moreover, this test has been already validated in different sets of high-risk asymptomatic individuals enrolled in CT-scan screening programmes [22]. In our study, the miR-Test showed a positive score in the majority of patients who subsequently were diagnosed with NSCLCs, with an accuracy of 71% (sensitivity 67%; specificity 72%). These values were slightly lower than those reported in the study by Montani et al (sensitivity and specificity of 77.8% and 74.8% respectively) [22]. These differences may be related to the small number of tumours observed in our study group that prevented any predictive value analysis. On the other hand, the routinely assessment of the miR-Test in different populations may strengthen the previous validated results. Indeed, a new chemoprevention trial has been already designed including the miR-Test as a predictive diagnostic biomarker [43]. Interestingly, the two patients with negative miR-Test scores showed positive telomerase activity in their sputum samples. This observation seems to suggest that the combined use of these two molecular biomarkers might be able to improve the risk assessment of NSCLC development in high-risk populations.
The usefulness of molecular genotyping of early-stage NSCLC remains controversial. Conflicting results have been reported on the predictive and prognostic role of the mutations identified in early-stage disease including EGFR and KRAS mutations [44–46]. However, molecular analysis of early-stage NSCLC may constitute the substrate not only for the implementation of personalised therapies but also for the identification of new diagnostic molecular biomarkers. In this study, all cases harboured somatic mutations in at least one cancer-related gene that likely represents the driver of these tumours. Notably, the patients were all current or former smokers and in four out of six tumours we identified somatic mutations of KRAS that were frequently associated with the carcinogenic effect of tobacco smoke [44]. Moreover, the incidence of KRAS aberrations was even higher than that described in advanced-stage NSCLC (67% versus 35%) [34–36].
Interestingly, no mutations were identified in EGFR gene that together with KRAS is the most commonly mutated gene in lung adenocarcinoma. Furthermore, we detected one rare somatic mutation of BRAF and three TP53 somatic mutations that have been previously described in nearly 45% of early-stage adenocarcinoma of high-risk patients [34–36]. Although a sample-size bias cannot be excluded, these significant inter-patient differences in gene mutation frequencies may be related to the peculiar clinical characteristics of the study group (i.e. high-risk individuals with current or former smoking history and lung nodules).
The small size of the invasive or minimally invasive adenocarcinomas of this series shows that genetic aberrations driving tumour progression develop very early in the tumourigenesis. This finding has been already observed in atypical adenomatous hyperplasia that represents the morphologic continuum ending in the full-blown adenocarcinoma. Previous studies have demonstrated that atypical adenomatous hyperplasia can harbour some of the genetic alterations found in adenocarcinomas, including mutations of KRAS, EGFR, and TP53 [47–49]. The presence of driver genetic events even at early stage of tumour development offers the opportunity to test these mutations in cfDNA as potential biomarkers for early detection of lung cancer. Recently, the study of Izumchenko et al provided the proof-of-concept that genetic alterations associated with very small early glandular neoplasms can be detected in paired circulating DNA even before they invade and acquire malignant potential [50]. Molecular profiling of cfDNA is emerging as key non-invasive tool for monitoring tumour progression and therapy response/resistance. However, concerns about the specificity and sensitivity of this assays in the screening setting have so far limited its clinical application [8, 51]. Indeed, the relatively high sensitivity of cfDNA hotspot mutation analysis tests (e.g. digital PCR) requires a prior knowledge of the tumour mutational profile which pose a major limitation of its use for screening purpose [52]. It should be noted, however, that next generation sequencing (NGS) technologies allow for simultaneous assessment of multiple genetic aberrations in ‘unsupervised’ manner [52, 53]. These high-throughput assays have recently shown the ability to detect cfDNA mutations even in early-stage lung tumours [52, 54, 55]. These pioneer results may pave the way for future application of NGS-based cfDNA analysis as potential biomarker for lung cancer screening.
Our study has several limitations, including the small sample size, which precluded statistical subgroup analyses. Indeed, given the specific primary endpoint of the NCT00321893 trial, the sample size was not powered to define the statistically significance of circulating biomarkers and early detection of lung cancer. In addition, this is a retrospective study performed on samples prospectively collected before the trial completion date. For this reason, sputum and blood samples at the time of cancer diagnosis were not available. However, this is an exploratory analysis of potential circulating biomarkers of lung cancer development in a unique prospective cohort of high-risk individuals enrolled in a lung cancer chemoprevention trial. The new era of screening studies will be grounded in multidisciplinary strategies including clinical information, imaging approaches, and circulating biomarkers analyses that might allow for non-invasive highly sensitive and specific early detection of lung cancer.
Conclusion
In conclusion, our exploratory study confirms the pivotal role of screening programmes and highlights the clinical value of circulating biomarkers, including miRNA and sputum telomerase activity tests, in detecting clinically-silent early-stage NSCLC. Moreover, we corroborate the notion that even clinically and histologically early lung adenocarcinoma may be underpinned by somatic genetic events similar to those described in advanced-stage NSCLCs.
Acknowledgments
Laboratory technicians of Division of Pathology, European Institute of Oncology, Milan, Italy; Eva Szabo, MD, Lung & Upper Aerodigestive Cancer Research Group Division of Cancer Prevention, NCI, NIH, Bethesda, MD, USA.
Funding
The NCT00321893 trial was supported by the National Cancer Institute Division of Cancer Prevention, contract N01-CN-035159 to the UT MD Anderson Early Phase Chemoprevention Consortium.
Financial disclosures
FB is an inventor on a patent application regarding a diagnostic serum miRNA signature cited herein.
References
1. Torre LA et al (2015) Global cancer statistics 2012 CA Cancer J Clin 65(2) 87–108 DOI: 10.3322/caac.21262 PMID: 25651787
2. Veronesi G et al (2014) Diagnostic performance of low-dose computed tomography screening for lung cancer over five years J Thorac Oncol 9(7) 935–9 DOI: 10.1097/JTO.0000000000000200 PMID: 24922008
3. http://seer.cancer.gov/statfacts/html/lungb.html in accessed 25 February 2016
4. Church TR et al (2013) Results of initial low-dose computed tomographic screening for lung cancer N Engl J Med 368(21) 1980–91 DOI: 10.1056/NEJMoa1209120 PMID: 23697514 PMCID: 3762603
5. Aberle DR et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening N Engl J Med 365(5) 395–409 DOI: 10.1056/NEJMoa1102873 PMID: 21714641 PMCID: 4356534
6. Veronesi G et al (2008) Difficulties encountered managing nodules detected during a computed tomography lung cancer screening program J Thorac Cardiovasc Surg 136(3) 611–617 DOI: 10.1016/j.jtcvs.2008.02.082 PMID: 18805261
7. Veronesi G et al (2016) The challenge of small lung nodules identified in CT screening: can biomarkers assist diagnosis? Biomark Med 10(2) 137–43 DOI: 10.2217/bmm.15.122 PMID: 26764294
8. Karampini E and McCaughan F (2016) Circulating DNA in solid organ cancers-analysis and clinical application QJM 109(4) 223–7 DOI: 10.1093/qjmed/hcv146
9. Esposito A et al (2014) Monitoring tumor–derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities Cancer Treat Rev 40(5) 648–55 DOI: 10.1016/j.ctrv.2013.10.003
10. Marzese DM, Hirose H and Hoon DS (2013) Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients Expert Rev Mol Diagn 3(8) 827–44 DOI: 10.1586/14737159.2013.845088
11. Sozzi G et al (2003) Quantification of free circulating DNA as a diagnostic marker in lung cancer J Clin Oncol 21(21) 3902–8 DOI: 10.1200/JCO.2003.02.006 PMID: 14507943
12. Szpechcinski A et al (2009) Real-time PCR quantification of plasma DNA in non-small cell lung cancer patients and healthy controls Eur J Med Res 14 Suppl 4 237–40 DOI: 10.1186/2047-783X-14-S4-237
13. Yoon KA et al (2009) Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls J Mol Diagn 11(3) 182–5 DOI: 10.2353/jmoldx.2009.080098 PMID: 19324991 PMCID: 2671334
14. Belinsky SA et al (2002) Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers Cancer Res 62(8) 2370–7 PMID: 11956099
15. Hashim M et al (2011) Prognostic significance of telomerase activity and some tumor markers in non-small cell lung cancer Med Oncol 28(1) 322–30 DOI: 10.1007/s12032-010-9444-0
16. Schramm M et al (2011) Equivocal cytology in lung cancer diagnosis: improvement of diagnostic accuracy using adjuvant multicolor FISH, DNA-image cytometry, and quantitative promoter hypermethylation analysis Cancer Cytopathol 119(3) 177–92 DOI: 10.1002/cncy.20142 PMID: 21413159
17. Chen M, Calin GA and Meng QH (2014) Circulating microRNAs as promising tumor biomarkers Adv Clin Chem 67 189–214 DOI: 10.1016/bs.acc.2014.09.007 PMID: 25735862
18. Bianchi F (2015) Lung cancer early detection: the role of circulating microRNAs EBioMedicine 2(10) 1278–9 DOI: 10.1016/j.ebiom.2015.08.032 PMID: 26629506 PMCID: 4634623
19. Sozzi G et al (2014) Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study J Clin Oncol 32(8) 768–73 DOI: 10.1200/JCO.2013.50.4357 PMID: 24419137 PMCID: 4876348
20. Wozniak MB et al (2015) Circulating microRNAs as non-Invasive biomarkers for early detection of non-small-cell lung cancer PLoS One 10(5) e0125026 DOI: 10.1371/journal.pone.0125026 PMID: 25965386 PMCID: 4428831
21. Bianchi F et al (2011) A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer EMBO Mol Med 3(8) 495–503 DOI: 10.1002/emmm.201100154 PMID: 21744498 PMCID: 3377091
22. Montani F et al (2015) miR-Test: a blood test for lung cancer early detection J Natl Cancer Inst 107(6) djv063 DOI: 10.1093/jnci/djv063 PMID: 25794889
23. Wang C et al (2015) A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients EBioMedicine 2(10) 1377–85 DOI: 10.1016/j.ebiom.2015.07.034 PMID: 26629532 PMCID: 4634198
24. Veronesi G et al (2011) Randomized phase II trial of inhaled budesonide versus placebo in high-risk individuals with CT screen-detected lung nodules Cancer Prev Res (Phila) 4(1) 34–42 DOI: 10.1158/1940-6207.CAPR-10-0182
25. Hochmuth F, Jochem M and Schlattmann P (2015) Meta-analysis of aspirin use and risk of lung cancer shows notable results Eur J Cancer Prev 25(4) 258–68
26. Mc Menamin Ú et al (2015) Low-dose aspirin and survival from lung cancer: a population-based cohort study BMC Cancer 15 911 DOI: 10.1186/s12885-015-1910-9 PMID: 26573580 PMCID: 4647502
27. Lazzeroni M et al (2010) Budesonide versus placebo in high-risk population with screen-detected lung nodules: rationale, design and methodology Contemp Clin Trials 31(6) 612–9 DOI: 10.1016/j.cct.2010.08.006 PMID: 20719253 PMCID: 2962433
28. Veronesi G et al (2015) Long-term effects of inhaled budesonide on screening-detected lung nodules Ann Oncol 26(5) 1025–30 DOI: 10.1093/annonc/mdv064 PMID: 25672894 PMCID: 4405280
29. Herman JG and Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation N Engl J Med 349(21) 2042–54 DOI: 10.1056/NEJMra023075 PMID: 14627790
30. Cirincione R et al (2006) Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study Int J Cancer 118(5) 1248–53 DOI: 10.1002/ijc.21473
31. Sozzi G et al (2009) Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study Am J Respir Crit Care Med 179(1) 69–74 DOI: 10.1164/rccm.200807-1068OC
32. Travis WD et al (2015) WHO classification of tumours of the lung, pleura, thymus and heart In. IV ed. Lyon: IARC
33. Fumagalli C et al (2014) Prevalence and clinicopathologic correlates of O6-methylguanine-DNA methyltransferase methylation status in patients with triple-negative breast cancer treated preoperatively by alkylating drugs Clin Breast Cancer 14(4) 285–90 DOI: 10.1016/j.clbc.2014.02.010 PMID: 24709436
34. Cerami E et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data Cancer Discov 2(5) 401–4 DOI: 10.1158/2159-8290.CD-12-0095 PMID: 22588877 PMCID: 3956037
35. Gao J et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal Sci Signal 6(269) pl1 DOI: 10.1126/scisignal.2004088 PMID: 23550210 PMCID: 4160307
36. Cancer Genome Atlas Research Network (2014) Comprehensive molecular profiling of lung adenocarcinoma Nature 511(7511) 543–50 DOI: 10.1038/nature13385
37. Veronesi G et al (2007) Role of positron emission tomography scanning in the management of lung nodules detected at baseline computed tomography screening Ann Thorac Surg 84(3) 959–65 discussion 965–56 DOI: 10.1016/j.athoracsur.2007.04.058 PMID: 17720408
38. Veronesi G et al (2013) Computed tomography screening for lung cancer: results of ten years of annual screening and validation of cosmos prediction model Lung Cancer 82(3) 426–30 DOI: 10.1016/j.lungcan.2013.08.026 PMID: 24099665
39. van Engeland M et al (2002) K-ras mutations and RASSF1A promoter methylation in colorectal cancer Oncogene 21(23) 3792–5 DOI: 10.1038/sj.onc.1205466 PMID: 12032847
40. Li J et al (2003) RASSF1A promoter methylation and Kras2 mutations in non small cell lung cancer. Neoplasia 5(4) 362–6 DOI: 10.1016/S1476-5586(03)80029-5 PMID: 14511407 PMCID: 1550336
41. Fernández-Marcelo T et al (2015) Telomere length and telomerase activity in non-small cell lung cancer prognosis: clinical usefulness of a specific telomere status J Exp Clin Cancer Res 34 78 DOI: 10.1186/s13046-015-0195-9 PMID: 26250468 PMCID: 4528384
42. Ilie M et al (2014) “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease PLoS One 9(10) e111597 DOI: 10.1371/journal.pone.0111597 PMID: 25360587 PMCID: 4216113
43. Veronesi G et al (2015) Chemoprevention studies within lung cancer screening programmes Ecancermedicalscience 9 597 DOI: 10.3332/ecancer.2015.597 PMID: 26635901 PMCID: 4664502
44. Shepherd FA et al (2013) Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy J Clin Oncol 31(17) 2173–81 DOI: 10.1200/JCO.2012.48.1390 PMID: 23630215 PMCID: 4881333
45. Villaruz LC et al (2013) The prognostic and predictive value of KRAS oncogene substitutions in lung adenocarcinoma Cancer 119(12) 2268–74 DOI: 10.1002/cncr.28039 PMID: 23526491 PMCID: 3674175
46. Azzoli CG (2015) Practical value of molecular pathology in stage I-III lung cancer: implications for the clinical surgeon Ann Surg Oncol 22(11) 3459–65 DOI: 10.1245/s10434-015-4704-z PMID: 26215190
47. Westra WH et al (1996) K-ras oncogene activation in atypical alveolar hyperplasias of the human lung Cancer Res 56(9) 2224–8 PMID: 8616876
48. Yoshida Y et al (2005) Mutations of the epidermal growth factor receptor gene in atypical adenomatous hyperplasia and bronchioloalveolar carcinoma of the lung Lung Cancer 50(1) 1–8 DOI: 10.1016/j.lungcan.2005.04.012 PMID: 15950315
49. Slebos RJ et al (1998) p53 alterations in atypical alveolar hyperplasia of the human lung. Hum Pathol 29(8) 801–8 DOI: 10.1016/S0046-8177(98)90448-8 PMID: 9712420
50. Izumchenko E et al (2015) Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA Nat Commun 6 8258 DOI: 10.1038/ncomms9258 PMID: 26374070 PMCID: 4595648
51. Atwater T and Massion PP (2016) Biomarkers of risk to develop lung cancer in the new screening era Ann Transl Med 4(8) 158 DOI: 10.21037/atm.2016.03.46 PMID: 27195276 PMCID: 4860477
52. Fernandez-Cuesta L et al (2016) Identification of circulating tumor DNA for the early detection of small-cell lung cancer EBioMedicine 10 117–23 DOI: 10.1016/j.ebiom.2016.06.032 PMID: 27377626 PMCID: 5036515
53. Bianchi F (2015) Molecular profile of liquid biopsies: next generation biomarkers to improve lung cancer treatment Ecancermedicalscience 9 598 DOI: 10.3332/ecancer.2015.598 PMID: 26635902 PMCID: 4664509
54. Jamal-Hanjani M et al (2016) Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer Ann Oncol 27(5) 862–7 DOI: 10.1093/annonc/mdw037 PMID: 26823523
55. Chen KZ et al (2016) Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing Sci Rep 6 31985 DOI: 10.1038/srep31985 PMID: 27555497 PMCID: 4995492