Presence of human papillomavirus in breast cancer and its association with prognostic factors
Andreína Fernandes1, Gino Bianchi2, Adriana Pesci Feltri3, Marihorgen Pérez2 and María Correnti1
1Laboratorio de Genética Molecular. Instituto de Oncología y Hematología, MPPS, Caracas 1050, Venezuela
2Sección de Patología Mamaria, Instituto de Anatomía Patológica José O’Daly, Universidad Central de Venezuela, Caracas 1050, Venezuela
3Servicio de Ginecología, Unidad de Patología Mamaria, Hospital Universitario de Caracas, MPPS, Caracas 1050, Venezuela
Correspondence to: Andreína Fernandes. Email: andreinafernandes@yahoo.es
Abstract
Breast cancer accounts for 16% of all female cancers worldwide, and in Venezuela, it is the leading cause of death among women. Recently, the presence of high-risk genotypes of human papillomavirus (HPV) has been demonstrated in breast cancer and has been associated with histopathological features of the tumours. In Venezuela, there is no study which determines the association between the presence of HPV in breast cancer and the histopathological features. The aim of this investigation is to evaluate the presence of HPV in the different types of breast cancer, according to their molecular classification, based on the expression of ER, PR, HER2 and Ki67. With this purpose in mind, we assessed the presence of the HPV genome in 24 breast cancer samples diagnosed with infiltrating ductal carcinoma, ductal carcinoma in situ (DCIS) and lobular carcinoma, by the INNO-LIPA genotyping extra kit and the evaluation of the markers ER, PR, HER2, and Ki67 by immunohistochemistry. The viral genome was found in 41.67% of the total number of samples, 51 being the most frequent genotype with 30.77%, followed by types 18 and 33, with 23.08%, respectively. Most tumours were found in the group of luminal A, with a low range of Ki67 expression. The presence of HPV in breast tumours could affect their growth pattern and metastatic power.
Keywords: breast cancer, human papillomavirus, prognostic factors, molecular subtypes
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 25/06/2015; Received: 04/03/2015
Background
Breast cancer is the most common cancer in women in both developed and developing countries, representing 16% of all female cancers. It is estimated that over 508,000 women died in 2011 worldwide due to breast cancer. Although breast cancer is thought to be a disease of the developed world, almost 50% of breast cancer cases and 58% of deaths occur in less developed countries [1]. It is estimated that 45% of the 1.35 million new cases diagnosed each year, and more than 55% of breast cancer-related deaths occur in low- and middle-income countries. An estimated 1.7 million women will be diagnosed with breast cancer in 2020, a 26% increase from current levels, mostly in the developing world [2].
In Venezuela, according to the Yearbook Mortality of the Ministry of Popular Power for Health (MPPS) for 2011, breast cancer was the leading cause of cancer death among women, followed by cervical cancer, reporting 1.942 deaths, which represent 17.8% of all cancer deaths [3].
Breast cancer is classified according to the tumour node metastasis (TNM)-staging system based on tumour size (T), the presence or absence of metastases in the lymph nodes (N), and the presence of distant metastases (M) (American Joint Committee Cancer, AJCC, 2010), thus allowing for tumour stages [4].
Since late in the last century, molecular biology has provided a new perspective that has led to a different therapeutic approach, allowing a better understanding of the clinical prognosis and predicting response to systemic treatments [5].
The expression of hormone receptors, oestrogen receptors (ER), and progesterone receptors (PR), as well as overexpression or amplification of the human epidermal growth factor receptor 2 (HER2), have been identified as important predictors for patients with breast cancer. Currently, these markers are commonly used to define treatment and establish disease prognosis associated with clinical and pathological variables; such as lymph node involvement, tumour size, histological type, tumour grade, and surgical margins [6].
Approximately, two-thirds of breast tumours express ER and PR activation in the centre of the tumour, making them candidates for hormonal therapy. Another 20% have overexpression of HER2. These tumours may benefit from therapy with trastuzumab, a monoclonal antibody, that can be used alone or combined with chemotherapy [6, 7].
Breast tumours are classified into four subtypes, depending on their immunohistochemical profiles and based on deoxyribonucleic acid (DNA) microarrays. They are as follows: (a) Luminal A: ER , PR , HER2−, (b) Luminal B: ER , PR , HER2 , (c) Triple negative: ER−, PR−, HER2−, (d) HER2: ER−, PR−, HER2 [8]. Luminal tumours have been associated with favourable prognosis with 80–85% survival at 5 years; while the triple negative and HER2 positive are associated with poor prognoses, with a high risk of recurrence in a period of 3 years and a high mortality rate in 5 years, with survival rate decreases to 50–60% [6, 8].
Despite advances in molecular techniques, the aetiological factor of breast cancer has not been determined. A risk factor that has been proposed for several years is the infection with human papillomavirus (HPV) in breast tissue, due to its high carcinogenic potential. This DNA virus expresses E6 and E7 oncogenes, which interact with p53 and pRB, respectively, facilitating the development of malignancies due to uncontrolled cell cycle activation and inhibition of apoptosis [9]. It is very common that the genome of the high-risk HPV types has been integrated into the host genome during carcinogenesis, which usually leads to a loss in the regulation of E6 and E7 transcription [10].
Reports on the distribution of the HPV infection in breast cancer are controversial. Several authors have found no relationship between the presence of HPV genome sequences and the development of breast cancer [11, 12, 13, 14, 15]. On the other hand, a moderate frequency of HPV infection in patients with breast cancer has been reported, ranging between 20–48%, while other authors reported a high frequency between 60–85% [16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29].
This evidence allows a plausible relationship between breast cancer and the HPV infection, and it suggests that the viral infection may be a possible risk factor in the development of the disease that currently causes so many deaths worldwide among the female population. In Venezuela, there are no studies that correlate the presence of HPV with the prognostic markers of breast cancer, which is why this study aims to evaluate the presence of human papillomavirus in different types of breast cancer, according to the molecular classification based on the ER, PR, HER2, and Ki67 expression.
Methods
Study subjects
The total sample for the study included 24 patients diagnosed with ductal carcinoma in situ (DCIS), infiltrating ductal carcinoma and lobular carcinoma, in different stages according to the TNM classification. Fresh biopsies were obtained from patients who underwent surgery between February 2011 and March 2013, in the Gynecology Service, Breast Pathology Unit, of the University Hospital of Caracas, and cut into two sections. One section was frozen at −70°C for molecular analysis, and the other fragment was fixed in 10% neutral formalin and was paraffin-embedded. Each patient underwent a survey to collect clinical and socio-cultural data, and each was asked to sign an informed consent. This protocol was approved by the Bioethics Committee of the University Hospital of Caracas.
DNA extraction and quality assessment of genetic material
To perform the DNA extraction from fresh breast biopsies, we used the QIAmp DNA Mini Kit (250) (QIAGEN. Hilden, Germany), following the manufacturer’s instructions. The quality of the isolated DNA was checked by polymerase chain reaction (PCR) of control genes with primers generating fragments of 100, 200, 300, 400, and 600 bp [30]. The reaction mixture was prepared using 0.4 μl of dNTPs (100 mM), 5 μl of each primer (100 pM), 6.5 μl of 10X buffer, 5 μl of MgCl2 (50 mM), 0.5 μl of Taq polymerase, and 27.6 μl of nuclease-free water, to a final volume of 50 μl. The amplification conditions were 7 min at 95°C, 35 cycles of 30 sec at 95°C, 40 sec at 60°C, and 40 sec at 70°C and a final amplification for 15 s at 70°C.
HPV detection and genotyping
Detection and genotyping of the HPV genome from fresh breast biopsies was performed using the INNO-LIPA HPV Genotyping Extra test (Innogenetics, Belgium), following the manufacturer’s instructions. This test is a line probe assay, based on the reverse hybridization principle, designed for the identification of 28 different genotypes of the HPV by detection of specific sequences in the L1 region of the HPV. The assay uses the proven SPF10 primer set for the highly sensitive amplification of most clinically relevant HPV genotypes.
Immunohistochemistry
The breast cancer biopsies were fixed in 10% buffered formalin and embedded in paraffin blocks. About 3 μm sections were obtained and placed on glass slides. The tissues were deparaffinized with xylene and rehydrated with serial dilutions of alcohol. Sections were incubated with primary antibody. The primary antibodies used were as follows: 1D5 to detect ER (1:50); and 1294 for PR (1:50) (Dako, Carpinteria, USA). The HercepTest (1:300) was used for the detection of HER2 and MIB-1 for Ki67 (1:100) (Dako, Carpinteria, USA). All antibodies were diluted in PBS. Then, a biotinylated anti-mouse secondary antibody (Dako Real EnVision, Denmark) was added, and the tissue sections were visualised by light microscopy. Appropriate controls were used.
Expression of Ki67 was evaluated according to the parameters published in Stathopoulos et al (2014) [31], where values ≤20% are considered low and values >20% are considered high.
Statistical analysis
For statistical analysis, a Chi square test was performed to assess the independence of the variables, with the IBM SPSS Statistics software, version 2.0. Values less than or equal to 0.05 were considered statistically significant.
Results
We evaluated the HPV presence in 24 samples of patients with breast cancer operated on at the University Hospital of Caracas, between the period of February 2011 and March 2013. The overall average age of patients with cancer was 56.75 ± 12.79 years (range: 37–84). The distribution of tumours according to the histological diagnosis was 83.33% for infiltrating ductal carcinomas and 8.33% for DCIS and lobular ductal carcinomas. The distribution, according to the tumour stages, were 54.16% for stage II, 25% for stage I, 12.5% for stage III and 8.33% for stage 0.
HPV detection was evaluated by molecular biology. We found 41.67% of the viral genome in breast tumours. The most frequent genotype was 51, followed by types 33 and 18, high oncogenic risk types, with a 30.77%, 23.08%, and 15.38%, respectively. The genotypes 6 and 11, low oncogenic risk types, were found in 15.38% each. 30% of the samples showed mixed infection. Most HPV-positive patients were within the age range of 51–60 years.
Table 1 presents the HPV frequency according to various clinicopathological features, including risk factors associated with the development of cervical cancer, which in turn, coincide with some associated factors with the development of breast cancer.
Table 1. Clinical and histopathological features of breast tumours, according to the presence of HPV.
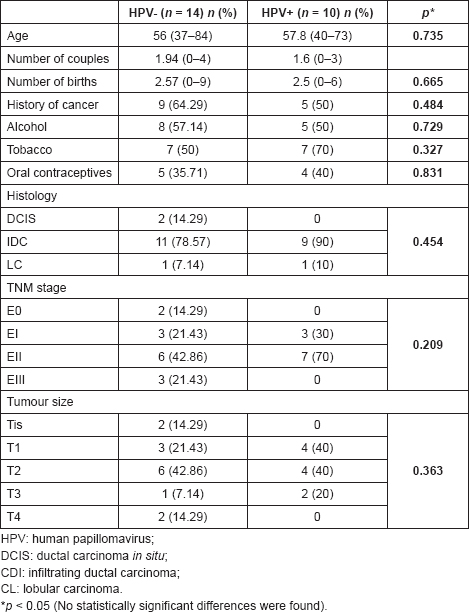
Table 2. Molecular subtypes of breast tumours, according to the presence of HPV.
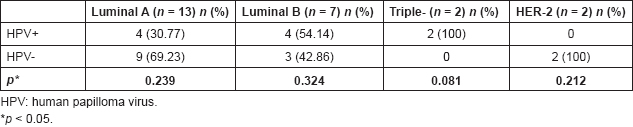
Breast tumours were classified based on the expression of the oestrogen and progesterone receptors, HER2 and the cell proliferation marker Ki67. Of the total sample, 54.17% was in the luminal A group, with an average value of 18% for Ki67 and 29. The luminal B group had 17%, with an average value of 15.71% for Ki67. The triple-negative group and HER2 were represented each by 8.33%, with an average value of 52.5% and 70% for Ki67, respectively.
According to this classification, the luminal A had 30.77% positivity for HPV, both low and high oncogenic risk in single and mixed infections. The luminal B group showed 57.14% positivity for HPV genotypes, equally low and high risk, but in single infections. In triple-negative patients, the presence of high oncogenic risk HPV was detected. The HER2 was the only group not associated with viral infection (Table 2). Although no statistically significant differences were found, the triple-negative group showed borderline significance.
Discussion
In this study, the presence of HPV was detected in 41.67% of breast cancer fresh biopsies. This detection rate is within the published data worldwide (10–86%) [16, 17, 18, 19, 20, 24, 25, 28], where the works of Antonsson et al (2011) [32] and Glenn et al (2012) [33] show a frequency of 50% each. In Latin America, the rate is a little lower. It is reporting between 5–40% of positivity [26, 27, 28, 34], particularly in the case of Herrera et al (2013) [34], who report a 40% of positivity. Their samples were from metaplastic breast tumours, a subset of breast tumours, characterised for being more aggressive and worse prognosis.
Li et al (2011) [35] and Simões et al (2012) [36] conducted an epidemiological study based on the inclusion of case–control and cross-sectional studies, prospective and retrospective, where they evaluated the presence of HPV in clinical breast cancer, using PCR assays. The overall prevalence of HPV DNA in patients with breast cancer was 24.49% (range: 0–86.21%) and 23% (range: 13.4–42.9%), respectively. South America studies showed a significant lower prevalence of HPV, 15.1% and 16.67%, respectively. Finally, all case–control studies were pooled, and the OR was calculated, which showed that HPV-positive women are 6.31 [35] and 5.9 [36] times more likely to have breast cancer, compared with HPV-negative women.
HPV genotypes found in our samples were mainly high oncogenic risk types, with 51, 33 and 18 being the most frequent. Similar to the studies of Akil et al (2008) [22], Heng et al (2009) [25], Antonsson et al (2011) [32] and Glenn et al (2012) [33] report a higher frequency of these HPV genotypes, most of them carried out with samples of Australian patients and another previous study done with samples of Venezuelan patients [37]. In the rest of the papers published worldwide, it is reported that the HPV genotype 16 is the most common [23]. Based on these reports, it can be suggested that the distribution of HPV infection in patients with breast cancer appears similar to that observed in the cervix, which also depends on factors such as geographic, ethnic and racial differences, and pattern reported previously by Correnti et al (2011) [38].
Despite this support, some studies found no evidence of HPV in any of their breast tissue samples [11, 12, 13, 14, 15]. These differences may be explained by variations in the methodological approaches used to detect HPV. Moreover, PCR protocols showing a diverse sensitivity and specificity are currently used to determine HPV presence in tumoural tissue and a ‘gold standard’ protocol has not been defined [26]. Also, the use of a different primer set could contribute to this issue [23].
In this study, the HPV detection was performed using the INNO-LIPA HPV Genotyping Extra test. This permits ultrasensitive detection of a broad spectrum of HPV genotypes by PCR with biotinylated primers SPF10 that amplifies a 65 bp long region in the HPV L1 gene, and typing by a reverse hybridization (LiPA), identifying 28 high- and low-risk genotypes [24].
It is worth noting that several authors have suggested that the presence of HPV is associated with smaller breast tumours [24, 27, 32, 34], when compared with negative tumours for viral infection. In this work, despite the fact that we found no statistical significant relationship, HPV-positive breast tumours were found mainly among the groups T1 and T2 lesions with a maximum size of 50 mm. According to Antonsson et al (2011) [32], this feature might imply that the presence of HPV could have an important role in the growth pattern and metastatic potential of breast carcinomas associated with a better prognosis, as in the case of tumours of the head and neck.
Regarding the molecular classification of breast cancer, samples from this study mainly corresponded to luminal group A, which have a high expression of oestrogen receptors and low levels of proliferation, resulting in low mitotic tumour cells and excellent prognosis [39], followed by luminal B tumours, triple negative, and HER2. This is consistent with those reported by Uribe et al (2010) [40], who reported a 60.63% rate of luminal A tumours in patients of Estado Lara, Venezuela. Stathopoulos et al (2014) [31] recommend using Ki67 as a predictive factor in luminal A type tumours, reporting values ≤20% in 99%. For luminal B, Ki67 distributed values found between low and high values, whereas in triple negative and HER2, Ki67 values were above 20%. The samples evaluated in our study had a similar behaviour, being able to establish a relationship between the molecular subtype and cell proliferation marker.
There are few studies that have evaluated the relationship between the presence of HPV and prognostic factors of breast cancer. Henning et al (1999) [17] and Damin et al (2004) [19] found no statistically significant differences between these variables. However, Kroupis et al (2006) [41] reported that HPV-positive tumours have lower positivity for RE, higher Ki67 expression and high histological grade. These authors indicated the need for further studies to evaluate the possibility that the proliferative potential and aggressive tumour phenotype is directly due to the presence of high-risk HPV at the time of carcinogenesis in the breast.
Currently, the mechanism of HPV infection in the breast is uncertain. Due to the fact that the HPV life cycle occurs in epithelial layers, bloodstream viraemia is not a possible event [34]. The following two hypotheses are considered: (1) infection via the nipple skin, as demonstrated in the work of DeVilliers et al (2005) [20], which indicate the presence of the HPV genome in the tumour tissue, as in the nipple, proposing a retrograde ductular pattern of viral spread. (2) Haematologic infection being shown by the presence of mononuclear cells carrying HPV, isolated from women with cervical cancer [42], HIV-infected children [43] and healthy donors [44]. Additionally, other authors reported the presence of HPV-16 in breast tumours from patients with a history of cervical lesions caused by this viral type [17, 45], which suggests that malignant transformation is the result of a cell transfection by the plasma flow from the primary tumour.
More recently, several studies have converged upon the innate immune DNA cytosine deaminase APOBEC3B (A3B) as a significant source of genomic uracil lesions and mutagenesis in multiple human cancers, including those of the breast, head, and neck, cervix, bladder, lung, ovary, and other tissues. This enzyme belongs to a protein family that has broad and overlapping functions in innate immunity by restricting viruses, transposons and other foreign DNA elements [46].
Vieira et al (2014) [46] showed that the upregulation of A3B mRNA is directly induced by the high-risk E6 HPV protein, transfecting the full-length HPV genome or a full-length HPV genome containing a stop codon within the E6 open reading frame in cell lines. Then, it was probed in samples of patients with head and neck cancer, where reported overexpression of A3B in HPV-positive tumours.
Ohba et al (2014) [47] conducted a similar work, but in the cell line MCF10A, to determine whether HPV can be the starting point of carcinogenesis in breast cancer, by overexpression of A3B, allowing excessive mutations, dysregulated cell cycle and subsequent transformation. Their results show that HPV infection induces overexpression of A3B and infected cells represent more malignant phenotype than parental cells. These malignant phenotypes were largely abrogated when A3B was knocked down in HPV-infected cells.
Conclusions
The HPV genome was found in 41.67% of all breast cancer samples, mainly identifying high-risk oncogenic genotypes. Most of the breast tumours were luminal A. There was no statistically significant difference between the presence of HPV and the prognostic factors. However, further studies involving a larger number of samples and the evaluation of other parameters are needed, such as viral load, integration of the viral oncogenes and several pathways for the introduction of genomic instability.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This study was funded by the Misión Ciencias project LPL-2007001088 and PEII project No. 2012001201. We thank Fatima Santos RT(T) for her help in manuscript preparation.
References
1. OMS Organización Mundial de la Salud [http://www.who.int/es/]. Accessed on: 20/06/2014
2. Tfayli A, Temraz S and Abou R et al (2010) Breast cancer in low- and middle-income countries: an emerging and challenging epidemic J Oncol DOI: 10.1155/2010/490631
3. MPPS—Ministerio del poder popular para la salud Anuario de mortalidad 2011 Available at: http://www.mpps.gob.ve. Accessed on: 20/06/2014
4. American joint committee on cancer Available at: http://www.cancerstaging.org. Accessed on: 23/08/2014
5. Rodríguez M, Lacruz J and Márquez M et al (2014) Factores pronósticos y predictivos en el cáncer de mama con sobre-expresión del HER-2/neu Rev Venez Oncol 26(3) 175–186
6. Duarte J, Bustamante M and Dinizza R et al (2011) Immunohistochemical profile and clinical-pathological variables in breast cancer Rev Assoc Med Bras 58(2) 178–187 DOI: 10.1016/S0104-4230(12)70178-1
7. Fiszman G and Jasnis M (2011) Molecular mechanisms of trastuzumab resistance in HER2 overexpressing breast cancer Int J Breast Cancer DOI: 10.4061/2011/352182
8. Sandhu R, Parker J and Jones W et al (2010) Microarray-based gene expression profiling for molecular classification of breast cancer and identification of new targets for therapy Lab Med 41(6) 364–372 DOI: 10.1309/LMLIK0VIE3CJK0WD
9. Münger K, Baldwin A and Edwards K et al (2004) Mechanisms of human papillomavirus-induced oncogenesis J Virol 78(21) 11451–11460 DOI: 10.1128/JVI.78.21.11451-11460.2004 PMID: 15479788 PMCID: 523272
10. Hooley E, Fairweather V and Clarke A et al (2006) The recognition of local DNA conformation by the human papillomavirus type 6 E2 protein Nucleic Acids Res 34(14) 3897–3908 DOI: 10.1093/nar/gkl466 PMID: 16914454 PMCID: 1557802
11. Wredel D, Luqmani Y and Coombes R et al (1992) Absence of HPV 16 and 18 DNA in breast cancer J Cancer 65 891–894 DOI: 10.1038/bjc.1992.186
12. Gopalkrishna V, Singh U and Sodhari P et al (1996) Absence of human papillomavirus DNA in breast cancer as revealed by polymerase chain reaction Breast Cancer Res Treat 39 197–202 DOI: 10.1007/BF01806186 PMID: 8872328
13. De Cremoux P, Thioux M and Lebigot I et al (2008) No evidence of human papillomavirus DNA sequences in invasive breast carcinoma Breast Cancer Res Treat 109 55–58 DOI: 10.1007/s10549-007-9626-4
14. Headu S, Kumar U and Hussain S et al (2011) Breast cancer and human papilomavirus infection: no evidence of HPV etiology of breast cancer in Indian women BMC Cancer 11 27 DOI: 10.1186/1471-2407-11-27
15. Silva R and Da Silva B (2011) No evidence for an association of human papillomavirus and breast carcinoma Breast Cancer Res Treat 125 261–264 DOI: 10.1007/s10549-010-1129-z
16. Di Lonardo A, Venuti A and Marcante ML (1992) Human papillomavirus in breast cancer Breast Cancer Res Treat 21 95–100 DOI: 10.1007/BF01836955 PMID: 1320958
17. Hennig E, Sou Z and Thoresen S et al (1999) Human papillomavirus 16 in breast cancers of women treated for high grade cervical intraepithelial neoplasia (CINIII) Breast Cancer Res Treat 53 121–135 DOI: 10.1023/A:1006162609420 PMID: 10326789
18. Yu Y, Morimoto T and Sasa M et al (2000) Human papillomavirus type 33 in breast cancers in Chinese Breast Cancer 7(1) 33–36 DOI: 10.1007/BF02967185 PMID: 11029768
19. Damin A, Kara R and Zettler C et al (2004) Evidence for an association of human papillomavirus and breast carcinomas Breast Cancer Res Treat 84(2) 131–137 DOI: 10.1023/B:BREA.0000018411.89667.0d PMID: 14999143
20. De Villiers E, Sandstrom R and zur Hausen H et al (2005) Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast Breast Cancer Res 7 R1–R11 DOI: 10.1186/bcr940 PMID: 15642157 PMCID: 1064094
21. Kan CY, Lacopetta B and Lawson J et al (2005) Identification of human papillomavirus DNA gene sequences in human breast cancer Br J Cancer 93 946–948 DOI: 10.1038/sj.bjc.6602778 PMID: 16222323 PMCID: 2361649
22. Akil N, Yasmeen A and Kassab A et al (2008) High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study Br J Cancer 99 404–407 DOI: 10.1038/sj.bjc.6604503 PMID: 18648363 PMCID: 2527786
23. Khan N, Castillo A and Koriyama C et al (2008) Human papillomavirus detected in female breast carcinomas in Japan Br J Cancer 99 408–414 DOI: 10.1038/sj.bjc.6604502 PMID: 18648364 PMCID: 2527789
24. Cantu de León D, Pérez Montiel D and Nemcova I et al (2009) Human papillomavirus (HPV) in breast tumors: prevalence in a group of Mexican patients BMC Cancer 9 26 DOI: 10.1186/1471-2407-9-26
25. Heng B, Glenn W and Ye Y et al (2009) Human papilloma virus is associated with breast cancer Br J Cancer 101 1345–1350 DOI: 10.1038/sj.bjc.6605282 PMID: 19724278 PMCID: 2737128
26. Aguayo F, Khan N, Koriyama C and González C et al (2011) Human papillomavirus and Epstein-Barr virus infections in breast cancer from Chile Infect Agent Cancer 6(1) 7 DOI: 10.1186/1750-9378-6-7 PMID: 21699721 PMCID: 3141534
27. Herrera R, Khan N and Koriyama C et al (2011) High-risk human papillomavirus in mammary gland carcinomas and non-neoplastic tissues of Mexican women: no evidence supporting a cause and effect relationship The Breast 20 184–189 DOI: 10.1016/j.breast.2010.11.006
28. Pereira A, Lorenzetti M and González R et al (2013) Presence of human papilloma virus in a series of breast carcinoma from Argentina Plos One 8(4) e61613 DOI: 10.1371/journal.pone.0061613
29. Manzouri L, Salehi R and Shariatpanahi S et al (2014) Prevalence of human papilloma virus among women with breast cancer since 2005–2009 in Isfahan Adv Biomed Res 3 75 DOI: 10.4103/2277-9175.125873 PMID: 24627883 PMCID: 3950802
30. Van Dongen J, Langerak A and Brüggemann M et al (2003) Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted cation BMH4 CT98-3936 Leukemia 17 2257–2317 DOI: 10.1038/sj.leu.2403202
31. Stathopoulos G, Malamos N and Markopoulos C et al (2014) The role of Ki-67 in the proliferation and prognosis of breast cancer molecular classification subtypes Anticancer Drugs 25 950–957 DOI: 10.1097/CAD.0000000000000123 PMID: 24949917 PMCID: 4162382
32. Antonsson A, Spurr T and Chen A et al (2011) High prevalence of human papillomaviruses in fresh frozen breast cancer samples J Med Virol 83 2157–2163 DOI: 10.1002/jmv.22223 PMID: 22012724
33. Glenn W, Heng B and Delprado W et al (2012) Epstein-Barr virus, human papillomavirus and mouse mammary tumour virus as multiple viruses in breast cancer Plos One 7(11) e48788 DOI: 10.1371/journal.pone.0048788 PMID: 23183846 PMCID: 3501510
34. Herrera R, Vela T and Carrillo A et al (2013) High-risk human papillomavirus (HPV) DNA sequences in metaplastic breast carcinomas of Mexican women BMC Cancer 13 445 DOI: 10.1186/1471-2407-13-445
35. Li N, Bi X and Zhang Y et al (2011) Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis Breast Cancer Res Treat 126(2) 515–520 DOI: 10.1007/s10549-010-1128-0 PMCID: 3164261
36. Simões P, Medeiros L and Duarte P et al (2012) Prevalence of human papillomavirus in breast cancer A systematic review Int J Gynecol Cancer 22(3) 343–347 DOI: 10.1097/IGC.0b013e31823c712e PMID: 22214962
37. Fernandes A, Pesci-Feltri A and García I et al (2015) Evaluación de la infección por virus de papilloma humano en pacientes con cáncer de mama Rev Venez Oncol 27(1) 22–29
38. Correnti M, Medina F and Cavazza M et al (2011) Human papillomavirus (HPV) type distribution in cervical carcinoma, low-grade, and high-grade squamous intraepithelial lesions in Venezuelan women Gynecol Oncol 121 527–531 DOI: 10.1016/j.ygyno.2011.02.003 PMID: 21421263
39. Cornejo K, Kandil D and Khan A et al (2014) Theranostic and molecular classification of breast cancer Arch Pathol Lab Med 138 44–56 DOI: 10.5858/arpa.2012-0442-RA PMID: 24377811
40. Uribe J, Hernández C and Menolascino F et al (2010) Clasificación molecular del cáncer de mama y su correlación clínica Rev Venez Oncol 22(2) 109–116
41. Kroupis C, Markou Am Vourlidis N and Dionyssiou-Asteriou A et al (2006) Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics Clin Biochem 39 727–731
42. Pao C, Hor J and Yang F et al (1997) Detection of human papillomavirus mRNA and cervical cancer cells in peripheral blood of cervical cancer patients with metastasis J Clin Oncol 15 1008–1012 PMID: 9060540
43. Bodaghi S, Wood L and Roby G et al (2005) Could human papillomaviruses be spread through blood? J Clin Microbiol 43 5428–5434 DOI: 10.1128/JCM.43.11.5428-5434.2005 PMID: 16272465 PMCID: 1287818
44. Chen A, Keleher A and Kedda M et al (2009) Human papillomavirus DNA detected in peripheral blood samples from healthy Australian male blood donors J Med Virol 81 1792–1796 DOI: 10.1002/jmv.21592 PMID: 19697401
45. Widschwendter A, Brunhuber T and Wiedemair A et al (2004) Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history J Clin Virol 31 292–297 DOI: 10.1016/j.jcv.2004.06.009 PMID: 15494272
46. Vieira V, Leonard B and White E et al (2014) Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B mBio 5(6) e02234–e02314 DOI: 10.1128/mBio.02234-14 PMID: 25538195 PMCID: 4278539
47. Ohba K, Ichiyama K and Yajima M et al (2014) In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction PLoS ONE 9(5) e97787 DOI: 10.1371/journal.pone.0097787 PMID: 24858917 PMCID: 4032256