E6/E7 mRNA testing for human papilloma virus-induced high-grade cervical intraepithelial disease (CIN2/CIN3): a promising perspective
Massimo Origoni1, Paolo Cristoforoni2, Guia Carminati1, Chiara Stefani1, Silvano Costa3, Maria Teresa Sandri4, Luciano Mariani5 and Mario Preti6
1Department of Gynaecology & Obstetrics, Vita Salute San Raffaele University, School of Medicine, Milano 20132, Italy
2Polo Oncologico Villa Montallegro, Genova 16145, Italy
3M.F. Toniolo Hospital, Bologna 40141, Italy
4Division of Laboratory Medicine, European Institute of Oncology, Milano 20141, Italy
5HPV-UNIT, Regina Elena National Cancer Institute, Roma 00144, Italy
6Unit of Preventive Gynaecology, European Institute of Oncology, Milano 20141, Italy
Correspondence to: Massimo Origoni. Email: massimo.origoni@hsr.it
Abstract
Since the introduction of biomolecular testing for the identification of high-risk human papillomavirus DNA (hrHPV-DNA) in cervical cancer preventive strategies, many interesting aspects have emerged in this field; firstly, HPV-DNA testing has been demonstrated to have better sensitivity than conventional cytology in several settings: screening, triage of ASC-US and in follow-up after treatment. Despite this, some limitations of these new technologies have also been underlined: the major issue is the low specificity of the tests, which cannot discriminate between regressive and progressive infections. Thus, recent research has moved the attention towards novel markers of progression that could more precisely detect cases at real risk of cancer development. In view of the fact that progression to cancer is dependable of the E6/E7 proteins integration and transforming action, the overexpression of E6/E7 transcripts has been seen as a valuable marker of this risk. This review aims to summarise the literature data on this topic and to provide a clear view of the emerging perspectives.
Keywords: human papillomavirus, HPV, E6, E7, mRNA, cervical cancer, HPV-DNA
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 29/04/2015; Received: 10/12/2014
Background
To reduce morbidity and mortality caused by cervical cancer, in the past 40 years, most developed countries have introduced cervical screening programmes based on cytological examination of cervical smears (Pap test). However, cervical cytology demonstrated a less than optimal sensitivity, and advances in the knowledge of the natural history of the disease have more recently suggested alternative approaches. Persistent infection with high-risk genotypes of the human papillomavirus (hrHPV) has been recognised as the necessary cause of cervical cancer [1]. This led initially to the adoption of molecular tests evaluating hrHPV-DNA presence for the triage of minimally abnormal Pap smears. More recently, evidence has accumulated which implies that a significant improvement of the effectiveness of cervical cancer screening can be achieved by using hrHPV-DNA testing as a primary screening tool. As a matter of fact, hrHPV-DNA testing has a better sensitivity for clinically relevant cervical lesions (cervical intraepithelial neoplasia 2 or worse, CIN2+) than cytology. Moreover, molecular testing proved to offer better protection against high-grade CIN and cancer than cytology in subsequent screening rounds, allowing extension of the screening intervals with favourable logistic and economic implications. Nonetheless, hrHPV infections are rather common in the general population and most of them are transient, spontaneously regressing in a 18–24 months’ period. In fact, the positive predictive value (PPV) of a positive hrHPV-DNA test is rather low (i.e., only a small proportion of the women who test positive for high-risk HPV-DNA will have CIN2+ lesions at the time of the test or will develop it in the next few years), and up to 10% of the screened population test positive and will consequently need some form of second-level triage. The research effort has therefore been focused on potentially alternative or supplementary testing methods capable to limit unnecessary follow-up procedures of women with clinically not relevant, transient hrHPV infections. So far, most of the national guidelines on cervical cancer prevention consider reflex cytology (i.e., the lecture of the cytological sample collected at the time of primary HPV-DNA testing but stored and not initially evaluated) a valuable triage tool for hrHPV-DNA-positive women. Nevertheless, up to 8% of hrHPV-positive women with normal cytology have or will develop in the subsequent years a CIN2+ lesion, and the previous knowledge of HPV-DNA status might influence the subjective reading of cytology. Therefore, there is a strong need for biomarkers that will allow risk-stratification of HPV-positive women with normal cytology or, alternatively, capable of replacing cytology as a triage tool. Considering that the progression to cervical malignancy requires the overexpression of the E6 and E7 genes of the integrated hrHPV genome, demonstration in cervical samples of hrHPV E6/E7 transcripts might be more specific than hrHPV-DNA testing alone for the detection of CIN2+ lesions. Nowadays, transcript analysis is feasible on cervical scrapings, because the introduction of liquid-based cytology has resulted in collection media that preserve RNA sufficiently to allow in vitro amplification and detection.
hrHPV E6/E7 mRNA detection
Currently, three commercial tests exist for detecting hrHPV E6/E7 messenger RNA (mRNA). The PreTect HPV-Proofer (NorChip AS, Klokkarstua, Norway) and the NucliSENS Easy Q HPV (bioMérieux) are based on the same technology, but are produced by different companies, with small differences in mRNA extraction protocol and data analysis [2]. These tests are nucleic acid sequence-based amplification (NASBA). The assay is an isothermal, RNA real-time amplification method that detects E6/E7 mRNA and perform genotyping of the five more oncogenic hrHPV types (16, 18, 31, 33, and 45) [3]. The NASBA amplification is based on primer extension and transcription by coordinated activities of three enzymes (RNase H, reverse transcriptase and RNA polymerase). The amplification reaction is isothermal and performs at 41°C; due to this relatively low temperature, contaminating DNA cannot be amplified. Real-time detection using molecular beacons is based on the measurement of the time-related detection of increase in a fluorescent signal. This signal is produced only after specific binding of the molecular beacon to an amplicon (specific HPV mRNA) (figure 1). The measurement is performed in combination with the amplification step (real time) [4]. Both tests received the CE-IVD mark. (figure 1) The APTIMA HPV Assay (Hologic, San Diego, CA) is a target amplification nucleic acid probe test for the qualitative detection of E6/E7 viral mRNA from 14 hrHPV types (16/18/31/33/35/39/45/51/52/56/58/59/66/68). The assay involves three main events in a single tube: target capture, target amplification by transcription-mediated amplification (TMA) and detection of the amplification products. TMA is a transcription-based nucleic acid amplification method that involves two enzymes, reverse transcriptase and RNA polymerase (figure 2). The reverse transcriptase is used to generate a DNA copy of the target mRNA sequence containing a promoter sequence for RNA polymerase. RNA polymerase produces multiple copies of RNA amplicon from the DNA copy template [5]. The detection of the amplicons is performed by the hybridisation protection assay (HPA) using single-stranded nucleic acid probes with chemiluminescent labels that are specifically complementary to the target amplicons. The selection reagent differentiates between hybridised and unhybridised probes by inactivating the label on the unhybridised probes. During the detection step, light emitted from the labelled RNA/DNA hybrids is measured as photon signals in a luminometre and are reported as relative light units (RLU) [6]. APTIMA assay could also perform genotyping for HPV 16 single and 18-45 in pool but requires a separate APTIMA 16, 18/45 genotype assay. The APTIMA HPV assay is a FDA and CE-IVD approved test.
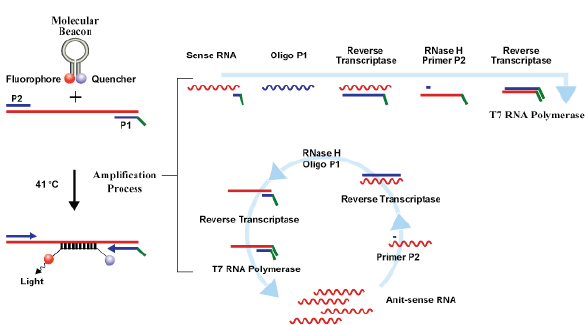
Figure 1. The NASBA RNA amplification technology.
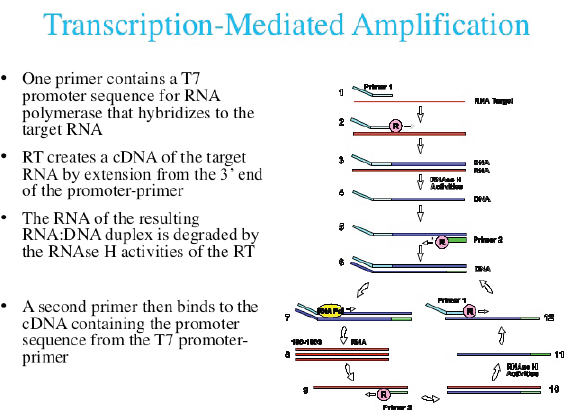
Figure 2. The TMA RNA amplification technology.
State of the art
In 2009, a group of researchers from Catholic University, Rome, published their findings on 180 colposcopically and histologically evaluated women, studied for the presence of HPV-DNA (HC II) and mRNA (NucliSENSE) [7]. As expected, fewer women tested positive for mRNA transcripts than hrHPV-DNA, and there was a fairly good correlation between the degree of positivity and the severity of the lesion. DNA from hrHPVs was found in 57.8% of the specimens; E6 and E7 transcripts were found in 45%. The rates of detection of HPV-DNA and of E6 and E7 transcripts were 33.3% and 25%, respectively, for specimens with normal findings; 51.4% and 31.9%, respectively, for specimens with cervical intraepithelial neoplasia grade 1 (CIN1); and 61.1% and 44.2% for specimens with CIN2, respectively. All specimens with CIN3 and 95.5% of specimens from patients with squamous cell carcinoma were positive by both assays. Overall, the mRNA tests showed a better specificity than the DNA tests for high-grade lesions (72.7% and 56.2%, respectively) and a higher positive predictive value (59.3% and 49.0%, respectively). The authors suggested that mRNA assays could be more powerful than DNA testing for predicting the risk of progression and offer a strong potential as a tool for triage and patient follow-up. In 2011, Burger from Norway performed a systematic review to determine the test performance of HPV mRNA testing compared to DNA testing using CIN2+ as the target condition [2]. The reference standard used to diagnose precancerous lesions was histologically confirmed cervical intraepithelial neoplasia 2+ (CIN2+). Sensitivity, specificity, positive and negative likelihood ratios and diagnostic odds ratios were evaluated for each study. The included studies (11 of 3271 publications) were of varying methodological quality and predominately performed in a secondary screening setting. Eight studies investigated the performance of the PreTect HPV-Proofer/NucliSENS EasyQ, two studies investigated the performance of the APTIMA assay, and one study investigated both mRNA tests on the same patient samples (Table 1). Due to few studies and considerable clinical heterogeneity, pooling of data was not possible; the authors, however, compiled a ‘best evidence synthesis’ for E6/E7 mRNA HPV testing. Sensitivities ranged from 0.41 to 0.86 and from 0.90 to 0.95 for the PreTect HPV-Proofer/Easy Q and APTIMA assay, respectively. Specificities ranged from 0.63 to 0.97 and from 0.42 to 0.61 for the PreTect HPV-Proofer/Easy Q and APTIMA assay, respectively. The SROC curves for both mRNA tests were to the left of the diagonal, and the APTIMA assay performed closest to the DNA tests. The authors concluded that mRNA tests have diagnostic relevance, but additional studies and economic evaluations have to be performed in order to make a solid conclusion regarding the clinical applicability of HPV mRNA testing. In 2012, Monsonego published the results from the FASE Study (French APTIMA screening evaluation), a multicentre trial involving 5,000 French women designed to assess the performance of APTIMA HPV assay (AHPV), hybrid capture 2 (HC2), in-house PCR genotyping, and ThinPrep LBC in population-based screening [8]. AHPV had the highest absolute risk of both histological endpoints, detecting 5% to 15% more CIN3+ and CIN2+ lesions, respectively, than LBC. Compared to the HC2 assay, the relative risk of AHPV positivity was 24% to 29% higher, with a significant difference in CIN2+ detection. With LBC as reference, AHPV had the best sensitivity/specificity balance measured by AUC (area under ROC curve) comparison test (significant for CIN2+), and the colposcopy referral rate (9.2%) comparable to that of LBC (8.7%). Data from the FASE study corroborate the suitability of AHPV in a primary cervical cancer screening. Several studies have investigated the PreTect HPV-Proofer assay on biopsy and cervical scraping samples and showed that the ratio of hrHPV E6/E7 mRNA positivity to hrHPV-DNA positivity increased along with the histological severity of dysplasia. This suggests a higher specificity of this mRNA assay for high-grade cervical lesions compared to HPV-DNA assays [9]. In a recent study by Rijkaart [10], 375 women among 13.401 women participating in a population-based cervical cancer screening programme in the Netherlands were stratified for CIN2+ risk according their hrHPV E6/E7 mRNA status. Study populations included 202 women with normal cytology, 88 with borderline or mild dyskaryosis (BMD) and 85 with moderate dyskaryosis or worse (>BMD); all of the women tested positive for hrHPV-DNA by the GP5+/6+ PCR assay used in the screening programme. A positive mRNA test result conferred an increased CIN2+ risk in hrHPV-DNA-positive women with normal cytology (0.55, 95% CI: 0.34–0.76) versus 0.20 (95% CI: 0.07–0.33) in mRNA-negative women. In hrHPV-DNA-positive women with BMD or >BMD, the result of the mRNA test did not influence the CIN2+ risk. Therefore, the authors concluded that mRNA testing by PreTect HPV-Proofer might be of value to select hrHPVDNA- positive women with normal cytology at an increased risk and therefore requiring immediate referral to colposcopy. A very recent study from Korea evaluated in a series of 337 ThinPrep samples a commercial diagnostic kit targeting a HPV E6/E7 mRNA based on RTqPCR assay and reported 91% sensitivity and 98.6% specificity for CIN2+ cervical lesions [11]. The overall positive rates of the HPV E6/ E7 mRNA RT-qPCR assay were 100% (24/24), 86% (38/44), 100% (7/7), 37% (10/27), 15% (5/32), and 1% (3/203) in SCC, HSIL, ASC-H, LSIL, ASC-US, and normal samples, respectively. The tested kit showed a remarkably higher sensitivity (91 vs. 41%) compared to the realtime NASBA assay, considered in the study as the reference standard. The tested assay showed also a better correlation between rate of positivity and severity of the cytological lesion and stressed the possible relevance of mRNA detection assays in the clinical management of women screened for cervical cancer precursors. In the discussion section, however, the authors underlined that the poor performance of the NASBA assay could be related to a series of technical limitations due to the adopted method of RNA extraction and amplification and stressed the importance of larger studies evaluating a more representative patient population as well as the importance of taking into account geographical differences, due to variable HPV genotypes prevalence in cervical cancer specimens. In summary, the performance in terms of sensitivity/specificity of the 5 hrHPV mRNA tests (PreTect HPV-Proofer and NucliSENS EasyQ HPV) cannot be pooled with the 14-h HPV mRNA test APTIMA. The APTIMA test is more similar to HPV DNA test like Hybrid Capture 2 regarding sensitivity and specificity, while the five genotype HPV E6/E7 mRNA tests have much higher specificity than the APTIMA assay [12, 13].
Table 1. Performance of E6/E7 HPV mRNA and DNA tests.
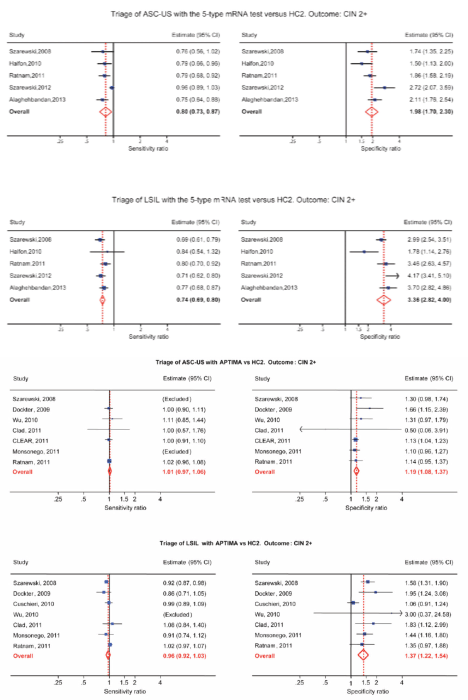
HPV mRNA testing in the triage of LSIL
HPV-DNA tests have low specificity and a high positivity rate in LSILs, making HPV-DNA test not particularly useful in triage of LSIL. The 14 genotype APTIMA test and especially the five genotype HPV E6/E7 mRNA tests have higher specificity and a low positivity rate in LSIL. A low positivity rate translates into a low referral rate for colposcopy, which is very appealing for triage situations [13–16]. Women with minor cervical lesions have a small but significantly increased risk of developing cervical cancer compared to women with normal smears. The purpose of triage is to identify women needing further follow-up by colposcopy and biopsy to detect high grade cervical dysplasia or cancer (CIN2+). In cases of atypical squamous cell of undetermined significance (ASC-US), it is now widely recognised that triage with an HPV-DNA test is more sensitive, but less specific than repeat cytology [17, 18]. For low-grade squamous intraepithelial lesions (LSIL), however, recommendations regarding the best triage method are conflicting and vary from repeat cytology, HPV testing or direct referral to colposcopy [19, 20]. Meta-analyses indicate that triage of LSIL with a high-risk HPV DNA test is not more sensitive and substantially less specific than repeat cytology [15, 16]. In some studies, HPV-based triage of LSIL in women older than 30–35 years has been suggested [21, 22], but other studies have not confirmed this [23, 24]. Defining the best strategy to triage LSIL lesions is, therefore, identified as a priority for research [25]. In a Norwegian study of 522 women with LSIL, 207 had biopsies and 125 of them had CIN2+. The sensitivity and specificity of repeat cytology (ASC-US or worse) were 85.7% (95% (CI): 72.1, 92.2) and 54.4 % (95% CI: 46.9, 61.9), respectively. The sensitivity and specificity of the HPV mRNA test were 94.2% (95% CI: 88.7, 99.7) and 86.0% (95% CI: 81.5, 90.5), respectively. The PPV of repeated cytology was 38.4% (95% CI: 29.9, 46.9) compared to 67.0% (95% CI: 57.7, 76.4) of the HPV mRNA test. In conclusion, five genotype HPV E6/E7 mRNA testings were more sensitive and specific than repeated cytology in triage of women with LSIL cytology. In addition, the HPV mRNA test showed higher PPV. These data indicate that the HPV mRNA test is a better triage test for women with LSIL than repeated cytology [15, 16]. In a meta-analysis by Arbyn, the pooled sensitivity and specificity of APTIMA to triage LSIL were 96.7% (95% CI: 91.4–98.9%) and 38.7% (95% CI: 30.5–47.6%) for CIN3+. APTIMA was as sensitive as HC2 but more specific (ratio: 1.35; 95% CI: 1.11–1.66). In both triage of ASC-US and LSIL, APTIMA is as sensitive but more specific than HC2 for detecting cervical pre-cancer [14]. In follow-up of patients with negative colposcopy/negative cervical histology, the five genotype HPV E6/E7 mRNA tests have a higher specificity and a higher PPV for CIN2+ than repeated cytology, suggesting a ‘test and treat’ approach for HPV mRNA-testing in women above 40 years [26]. In clinical studies, histology is used as a gold standard for detecting CIN2+. Unfortunately, colposcopy does not have optimal sensitivity for CIN2+. The National Health Service Cervical Screening Programme (NHSCSP) Guidelines for Colposcopy and Programme Management, which guides British practice, asks for evidence of a colposcopic accuracy of 65% [27]. Zuchna, reported 66.2% sensitivity of CIN2+ when up to three guided cervical biopsies were taken regarded as a diagnostic test with the cone specimen as reference standard [28]. Using digitised cervical images from 919 women referred for equivocal or minor cytologic abnormalities into the ASCUS-LSIL Triage Study, Massad, reported 39% sensitivity for CIN2+ [29]. Hence, all women with negative colposcopy and biopsies after abnormal cytology and/or HPV testing have to be followed.
HPV mRNA testing in the follow-up after conservative treatment for CIN2/CIN3 lesions
Women conservatively treated for CIN2–3 lesions are considered at high risk of developing invasive carcinoma for many years, and therefore, they require continued surveillance. In particular, between 5% and 20% of women treated for CIN2+ will develop recurrent disease within 3 years [30]. For this reason, an adequate follow-up is mandatory. Currently, despite the now perceived low sensitivity, Pap test is widely used in the follow-up of patients treated for intraepithelial neoplasia and European guidelines for cervical screening policy recommend 6-, 12- and 24-month cytology after CIN treatment [31, 32]. Observational studies demonstrated that HPV-DNA testing has a significant higher efficacy compared to cytology in predicting persistent disease or relapse, and hence, the detection of a persistent infection with hrHPV genotypes has the potential to improve patient management. In point of fact hrHPV-DNA testing is significantly more sensitive (90%) compared with follow-up cytology (70%) [33, 34] and currently ASCCP recommends the use of HPV-DNA testing with cytology (co-testing) at 12 and 24 months following treatment for CIN2/3 [35]. Actually, eradication of the clinical lesion does not necessarily mean eradication of all the infected tissue and up 40% of treated women still remains hr-HPV positive due to the fact that hrHPV-DNA testing does not distinguish between a persistent infection from a new, transient one [36, 37]. This is the reason why hrHPV-DNA testing owns a lower specificity than cytology in identifying relapse or persistent disease. Recent prospective studies have documented that rates of residual or recurrent disease in women with persistent hrHPV 16 and/or 18 are higher than in women with other hrHPV types, stressing the importance of type–specific genotyping determination after treatment for CIN2+ [38–40]. As hrHPV-genotyping assays have become increasingly utilised in clinical practice, further research will be needed to determine whether type-specific HPV results can improve the identification of women most likely at higher risk of developing cervical disease after treatment. A recent systematic review highlights that new HPV infections are a potential source for future disease risk after treatment for cervical pre-cancer and cancer [41]. More recently, it has been suggested that the oncogene activity by hr-mRNA transcripts [42] may be a better indicator of women at risk of persistence or relapse of the disease, showing an higher specificity than DNA-based assays, having a better specificity and negative predicting value (NPV) than hrHPV-DNA testing [43], while others underline that HPV E6/E7 mRNA is not useful to detect relapse [2]. In order to assess the performance of different biomolecular tests, alone or in cotesting with cytology, an Italian prospective study is on-going to evaluate the prediction of persistence/recurrence rate in women treated for CIN2+. Preliminary results seem to indicate that cotesting Pap + HPV E6/E7 mRNA shows the best specificity, while hrHPV genotyping retains the higher prediction for CIN2+ persistent or recurrent disease.
Conclusions
In general, HPV E6/E7 mRNA testing has been demonstrated in several published experiences to have the characteristic of an accurate diagnostic tool in the field of cervical cancer prevention [44]. For this reason, current evidence suggests that mRNA testing may be regarded with promising possibilities in terms of implementation of the diagnostic accuracy of pre-neoplastic cervical lesions in different settings [45–50], including glandular lesions and adenocarcinoma [51]. Primary screening has obviously been investigated more widely, representing the most interesting application option in view of the recent evidence-based shift from cytology to molecular-based screening programmes worldwide. Compared to hrHPV-DNA testing, which actually represents the most validated alternative to cytology in screening settings, mRNA tests present the valuable improvement of a better specificity, and consequently, higher positive predictive value (PPV) towards highgrade cervical lesions (CIN2+) [52]. In terms of test of cure after treatment, though with the significant limitation of a lack of large controlled trials, interesting preliminary data indicate a possible integration of mRNA testing with cytology. HPV E6/E7 mRNA testing may serve as a more specific discriminator between transient cervical dysplasia and potentially progressive lesions. Accordingly, testing for high-risk HPV E6/E7 mRNA might reduce the psychological burden associated with HPV-DNA testing [53].
In conclusion, since the beginning of the molecular era in the prevention of cervical cancer, many steps forward have already been made to overcome the limitations of cervical cytology alone; mRNA testing actually represents a new challenge, with the promising possibility of being integrated in the pool of valuable molecular tools that will hopefully lead to the elimination of invasive cervical cancer in women in the near future.
Acknowledgments
The authors wish to acknowledge the guidance and mentorship of Mario Sideri in this work and the entire activity of the Italian HPV Study Group (IHSG). He has been among the most intelligent and visionary colleagues we ever encountered and we were blessed with his friendship. The ‘Mario Sideri Italian HPV Study Group - IHSG’ endeavours to continue its research and teaching initiatives in his memory.
References
1. Tommasino M (2014) The human papillomavirus family and its role in carcinogenesis Semin Cancer Biol 26 13–21 DOI: 10.1016/j.semcancer.2013.11.002
2. Burger EA et al (2011) HPV mRNA tests for the detection of cervical intraepithelial neoplasia: a systematic review Gynecol Oncol 120 430–438 DOI: 10.1016/j.ygyno.2010.11.013
3. Jeantet D et al (2009) NucliSENS EasyQ HPV v1 test—testing for oncogenic activity of human papillomaviruses J Clin Virol 45 (Suppl 1) S29–S37 DOI: 10.1016/S1386-6532(09)70006-X
4. Deiman B, Van Aarle P and Sillekens P (2002) Characteristics and applications of nucleic acid sequence-based amplification (NASBA) Mol Biotechnol 20 163–179 DOI: 10.1385/MB:20:2:163 PMID: 11876473
5. Kacian DL and Fultz TJ Nucleic acid sequence amplification methods U.S. Patent 1995, 5,399, 491
6. Arnold LJ et al (1989) Assay formats involving acridinium-ester-labeled DNA probes Clin Chem 35 1588–1594 PMID: 2667804
7. Cattani P et al (2009) Clinical performance of human papillomavirus E6 and E7 mRNA testing for high-grade lesions of the cervix J Clin Microbiol 47 3895–3901 DOI: 10.1128/JCM.01275-09 PMID: 19828739 PMCID: 2786669
8. Monsonego J et al (2012) Risk assessment and clinical impact of liquid-based cytology, oncogenic human papillomavirus (HPV) DNA and mRNA testing in primary cervical cancer screening (The FASE Study) Gynecol Oncol 125 175–180 DOI: 10.1016/j.ygyno.2012.01.002 PMID: 22233689
9. Ratnam S et al (2010) Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancer J Clin Microbiol 48(8) 2779–2785 DOI: 10.1128/JCM.00382-10 PMID: 20573862 PMCID: 2916571
10. Rijkaart DC et al (2012) High-risk human papillomavirus (hrHPV) E6/E7 mRNA testing by PreTect HPV-proofer for detection of cervical high-grade intraepithelial neoplasia and cancer among hrHPV DNA-positive women with normal cytology J Clin Microbiol 50 2390–2396 DOI: 10.1128/JCM.06587-11 PMID: 22553244 PMCID: 3405578
11. Munkhdelger J et al (2014) Performance of HPV E6/E7 mRNA RT-qPCR for screening and diagnosis of cervical cancer with ThinPrep® Pap test samples Exp Mol Pathol 97 279–284 DOI: 10.1016/j.yexmp.2014.08.004 PMID: 25102300
12. Verdoodt F et al (2013) Triage of women with minor abnormal cervical cytology: meta-analysis of the accuracy of an assay targeting messenger ribonucleic acid of 5 high-risk human papillomavirus types Cancer Cytopathol 121(12) 675–687 DOI: 10.1002/cncy.21325 PMID: 23881840
13. Arbyn M et al (2013) The APTIMA HPV assay versus the Hybrid Capture 2 test in triage of women with ASC-US or LSIL cervical cytology: a meta-analysis of the diagnostic accuracy Int J Cancer 132(1) 101–118 DOI: 10.1002/ijc.27636
14. Sorbye SW et al (2010) Triage of women with minor cervical lesions: data suggesting a “test and treat” approach for HPV E6/E7 mRNA testing PLoS One 5(9) e12724 DOI: 10.1371/journal.pone.0012724
15. Sorbye SW et al (2011) Triage of women with low-grade cervical lesions–HPV mRNA testing versus repeat cytology PLoS One 6(8) e24083 DOI: 10.1371/journal.pone.0024083 PMID: 21918682 PMCID: 3168878
16. Sorbye SW et al (2014) HPV mRNA is more specific than HPV DNA in triage of women with minor cervical lesions PLoS One 9(11) e112934 DOI: 10.1371/journal.pone.0112934 PMID: 25405981 PMCID: 4236101
17. Arbyn M et al (2004) Virologic versus cytologic triage of women with equivocal Pap-smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia J Natl Cancer Inst 96 280–293 DOI: 10.1093/jnci/djh037 PMID: 14970277
18. Arbyn M et al (2005) Clinical utility of HPV-DNA detection: triage of minor cervical lesions, follow up of women treated for high-grade CIN: an update of pooled evidence Gynecol Oncol 99(3 Suppl 1) S7–11 DOI: 10.1016/j.ygyno.2005.07.033 PMID: 16154623
19. Boardman LA and Kennedy CM (2008) Management of atypical squamous cells, low-grade squamous intraepithelial lesions, and cervical intraepithelial neoplasia 1 Obstet Gyn Clin North Am 35 599–614 DOI: 10.1016/j.ogc.2008.09.001
20. Wright TC, Jr et al (2007) 2006 consensus guidelines for the management of women with abnormal cervical cancer screening tests Am J Obstet Gynecol 197 346–355 DOI: 10.1016/j.ajog.2007.07.047 PMID: 17904957
21. Ronco G et al (2007) HPV triage for low grade (L-SIL) cytology is appropriate for women over 35 in mass cervical cancer screening using liquid based cytology Eur J Cancer 43 476–480 DOI: 10.1016/j.ejca.2006.11.013 PMID: 17223540
22. Thrall MJ, Smith DA and Mody DR (2010) Women ≥ 30 years of age with low grade squamous intraepithelial lesion (LSIL) have low positivity rates when cotested for high-risk human Papillomavirus: should we reconsider HPV triage for LSIL in older women? Diagn Cytopathol 38 407–412
23. Arbyn M et al (2009) Triage of women with equivocal or low-grade cervical cytology results: a meta-analysis of the HPV test positivity rate J Cell Mol Med 13 648–659 DOI: 10.1111/j.1582-4934.2008.00631.x PMID: 19166485
24. Arbyn M, Martin-Hirsch P and Buntinx F (2010) Immediate colposcopy referral in women with low-grade abnormal results on cervical cytology detects more CIN2 or worse lesions than cytological surveillance in primary care, but might lead to overtreatment Evid Based Med 15 13–14 DOI: 10.1136/ebm.15.1.13 PMID: 20176871
25. Ovestad IT et al (2011) Comparison of different commercial methods for HPV detection in follow-up cytology after ASCUS/LSIL, prediction of CIN2-3 in follow up biopsies and spontaneous regression of CIN2-3 Gynecol Oncol 123(2) 278–283 DOI: 10.1016/j.ygyno.2011.07.024 PMID: 21835440
26. Sorbye SW et al (2011) HPV E6/E7 mRNA testing is more specific than cytology in post-colposcopy follow-up of women with negative cervical biopsy PLoS One 6(10) e26022 DOI: 10.1371/journal.pone.0026022 PMID: 21998748 PMCID: 3188582
27. The national health service cervical screening programme (NHSCSP) guidelines for colposcopy and programme management–second edition NHSCSP Publication No 20, May 2010. ISBN 978-1-84463-069-1 Available at: www.cancerscreening.nhs.uk
28. Zuchna C et al (2010) Diagnostic accuracy of guided cervical biopsies: a prospective multicenter study comparing the histopathology of simultaneous biopsy and cone specimen Am J Obstet Gynecol 203 321 e1–e6 DOI: 10.1016/j.ajog.2010.05.033
29. Massad LS et al (2009) National institutes of health/American society for colposcopy and cervical pathology (the NIH/ASCCP) research group. The accuracy of colposcopic grading for detection of high-grade cervical Intraepithelial neoplasia J Low Genit Tract Dis 13(3) 137–144 DOI: 10.1097/LGT.0b013e31819308d4 PMID: 19550210 PMCID: 2921444
30. Bollen LMJ et al (1999) Prediction of recurrent cervical dysplasia by human papillomavirus detection among patients with abnormal cytology Gynecol Oncol 72 199–201 DOI: 10.1006/gyno.1998.5250 PMID: 10021301
31. Paraskevaidis E et al (2004) The role of HPV DNA testing in the follow-up period after treatment for CIN: a systematic review of the literature Cancer Treat Rev 30 205–211 DOI: 10.1016/j.ctrv.2003.07.008 PMID: 15023438
32. Jordan J et al (2009) European guidelines for clinical management of abnormal cervical cytology, Part 2 Cytopathology 20 5–16 DOI: 10.1111/j.1365-2303.2008.00636.x PMID: 19133067
33. Soutter WP, Sasieni P and Paraskevaidis E (2006) Long-term risk of invasive cervical cancer after treatment of squamous cervical intraepithelial neoplasia Int J Cancer 118 2048–2055 DOI: 10.1002/ijc.21604
34. Heymans J et al (2011) Type specific HPV geno-typing improves detection of recurrent high-grade cervical neoplasia after conisation Int J Cancer 129 903–909 DOI: 10.1002/ijc.25745
35. Massad LS et al (2013) 2012 ASCCP consensus guidelines conference. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors Obstet Gynecol 121 829–846 DOI: 10.1097/AOG.0b013e3182883a34 PMID: 23635684
36. Costa S et al (2003) Factors predicting human papillomavirus clearance in cervical intraepithelial neoplasia lesions treated by conisation Gynecol Oncol 90 358–365 DOI: 10.1016/S0090-8258(03)00268-3 PMID: 12893200
37. Kocken M et al (2011) Risk of recurrent high-grade cervical intraepithelial neoplasia after successful treatment: a long-term multi-cohort study Lancet Oncol 12 441–450 DOI: 10.1016/S1470-2045(11)70078-X PMID: 21530398
38. Venturoli S et al (2008) Correlation of high-risk human papillomavirus genotypes persistence and risk of residual or recurrent cervical disease after surgical treatment J Med Virol 80 1434–1440 DOI: 10.1002/jmv.21198 PMID: 18551620
39. Zappacosta R et al (2013) Detection of residual/recurrent cervical disease after successful LEEP conization: the possible role of mRNA-HPV test Curr Pharm Des 19(8) 1450–1457
40. Frega A et al (2014) Assessment of HPV-mRNA test to predict recurrent disease in patients previously treated for CIN 2/3 J Clin Virol 60(1) 39–43 DOI: 10.1016/j.jcv.2014.01.017 PMID: 24602516
41. Rositch AF et al (2014) The incidence of human papillomavirus infection following treatment for cervical neoplasia: a systematic review Gynecol Oncol 132 767–779 DOI: 10.1016/j.ygyno.2013.12.040 PMID: 24412508
42. Cricca M et al (2007) Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions Gynecol Oncol 106 549–557 DOI: 10.1016/j.ygyno.2007.05.004 PMID: 17568661
43. Frega A et al (2014) Assessment of HPV-mRNA test to predict recurrent disease in patients previously treated for CIN 2/3 J Clin Virol 60 39–43 DOI: 10.1016/j.jcv.2014.01.017 PMID: 24602516
44. Oliveira A, Verdasca N and Pista Â. (2013) Use of the NucliSENS EasyQ HPV assay in the management of cervical intraepithelial neoplasia J Med Virol 85(7) 1235–1241 DOI: 10.1002/jmv.23590 PMID: 23918542
45. Benevolo M et al (2011) Sensitivity, specificity, and clinical value of human papillomavirus (HPV) E6/E7 mRNA assay as a triage test for cervical cytology and HPV DNA test J Clin Microbiol 49(7) 2643–2650 DOI: 10.1128/JCM.02570-10 PMID: 21525231 PMCID: 3147832
46. Zappacosta R et al (2015) Role of E6/E7 mRNA test in the diagnostic algorithm of HPV-positive patients showing ASCUS and LSIL: clinical and economic implications in a publicly financed healthcare system Expert Rev Mol Diagn 15(1) 137–150 DOI: 10.1586/14737159.2015.961915
47. Koliopoulos G et al (2012) The diagnostic accuracy of two methods for E6 & 7 mRNA detection in women with minor cytological abnormalities Acta Obstet Gynecol Scand 91(7) 794–801 DOI: 10.1111/j.1600-0412.2012.01414.x PMID: 22486415
48. Rijkaart DC et al (2012) High-risk human papillomavirus (hrHPV) E6/E7 mRNA testing by PreTect HPV-Proofer for detection of cervical high-grade intraepithelial neoplasia and cancer among hrHPV DNA-positive women with normal cytology J Clin Microbiol 50(7) 2390–2396 DOI: 10.1128/JCM.06587-11 PMID: 22553244 PMCID: 3405578
49. Munkhdelger J et al (2014) Comparison of the performance of the NucliSENS EasyQ HPV E6/E7 mRNA assay and HPV DNA chip for testing squamous cell lesions of the uterine cervix Diagn Microbiol Infect Dis 79(4) 422–427 DOI: 10.1016/j.diagmicrobio.2014.04.004 PMID: 24856365
50. Perez Castro S et al (2013) Human papillomavirus (HPV) E6/E7 mRNA as a triage test after detection of HPV 16 and HPV 18 DNA J Med Virol 85(6) 1063–1068 DOI: 10.1002/jmv.23544 PMID: 23588733
51. Hovland S et al (2010) E6/E7 mRNA expression analysis: a test for the objective assessment of cervical adenocarcinoma in clinical prognostic procedure Int J Oncol 36(6) 1533–1539 PMID: 20428778
52. Argyri E et al (2013) E6/E7 mRNA expression of high-risk HPV types in 849 Greek women Anticancer Res 33(9) 4007–4011 PMID: 24023342
53. Möckel J et al (2011) Human papillomavirus E6/E7 mRNA testing has higher specificity than liquid-based DNA testing in the evaluation of cervical intraepithelial neoplasia Anal Quant Cytol Histol 33(6) 311–315