p-Medicine: From data sharing and integration via VPH models to personalized medicine
S Rossi1, ML Christ-Neumann2, S Rüping2, FM Buffa3, D Wegener2, G McVie4, PV Coveney5, N Graf6 and M Delorenzi1
1Swiss Institute for Bioinformatics, Lausanne, Switzerland
2Fraunhofer IAIS, Schloss Birlinghoven, Sankt Augustin, Germany
3Weatherall Institute of Molecular Medicine, University of Oxford, Oxford, UK
4Ecancermedicalscience, Gotthardstrasse 20, 6300 Zug, Switzerland
5Centre for Computational Science, University College London, London, UK
6Department of Pediatric Oncology and Hematology, Saarland University, Homburg, Germany
Correspondence to: S Rossi. Email: simona.rossi@isb-sib.ch (or) N Graf. Email: graf@uks.eu
Abstract
The Worldwide innovative Networking in personalized cancer medicine (WIN) initiated by the Institute Gustave Roussy (France) and The University of Texas MD Anderson Cancer Center (USA) has dedicated its 3rd symposium (Paris, 6–8 July 2011) to discussion on gateways to increase the efficacy of cancer diagnostics and therapeutics (http://www.winconsortium.org/symposium.html).
Speakers ranged from clinical oncologist to researchers, industrial partners, and tools developers; a famous patient was present: Janelle Hail, a 30-year breast cancer survivor, founder and CEO of the National Breast Cancer Foundation, Inc. (NBCF).
The p-medicine consortium found this venue a perfect occasion to present a poster about its activities that are in accordance with the take home message of the symposium.
In this communication, we summarize what we presented with particular attention to the interaction between the symposium's topic and content and our project.
The concept
The knowledge about protein coding genes (PCGs), noncoding-RNAs (nc-RNAs), single-nucleotide polymorphisms (SNPs), and their behaviour in the human organism [1–3], which is being obtained from the analysis of data generated by array-based and DNA sequencing technologies, is impressive and is rapidly improving our understanding of the biology of cancer. Moreover, new tools have been developed to predict disease recurrence and progression, response to treatment, as well as new insights into various oncogenic pathways [4–7]. We expect that this will have a strong impact also on treatment decision-making in oncology over the next two decades.
Based on these advances, medicine is undergoing a revolution that is even transforming the nature of health care from reactive to proactive [8] according with the P4 concept that envisages the medicine as a predictive, personalized, preventive, and participatory discipline [9,10], or, in other words, a medicine that will use personal genomic information and other shared data to stratify the diseases in order to give specific treatments and ultimately stop disorder progression.
Expanding the P4 concept, Gorini and Pravettoni [11] indicated a fifth aspect, the psycho-cognitive, in order to give relevance to the behavioural component of the individuals (patients), foster their education and training in interacting with infrastructure like the one p-medicine will provide, help them to understand the medical documentation and to finally make well-informed choices (patient empowerment).
The p-medicine (www.p-medicine.eu) consortium is creating a biomedical platform to facilitate the translation from current practice to P5 medicine (predictive, personalized, preventive, participatory and psycho-cognitive) by integrating VPH models, clinical practice, imaging and omics data, in order to
p-medicine works closely together with the Virtual Physiological Human Network of Excellence (VPH–NoE). The VPH–NoE (http://www.vph-noe.eu/) is designed to foster and integrate pan-European research in the field of patient-specific computer models for personalized and predictive health care. Our emphasis is on formulating an open, modular framework of tools and services, including efficient secure sharing and handling of large personalized data sets, performing simulations of disease progression, building tools and models for VPH research [12,13], enrich, evaluate and validate results using literature mining.
p-Medicine clinical trials
To evaluate and validate the p-medicine infrastructure and its usability based on several categories of users (clinicians, patients, bioinformaticians, statisticians, biologists, data managers and tools developers), retrospective and prospective pilot clinico-genomic trials have been selected:
Role of SIB-BCF in p-medicine
SIB's Bioinformatics Core Facility (SIB-BCF) is providing evaluation guidelines and validation scenarios for p-medicine, based on its experience in low- and high-level data analysis (normalization, regression, survival analysis, annotation and meta-analysis) heterogeneous data management and experience gained during the ACGT project (http://www.eu-acgt.org/) in the field of quality assurance. SIB-BCF is identifying the objectives that need to be specifically tested (software components and services) in the context of data sources access, patient empowerment tools and clinical trial management.
The evaluation process will be performed in the context of scenarios that can be user group specific, common to several user groups, divided into sub-scenarios.
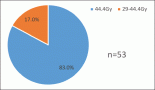
The p-medicine consortium is composed of heterogeneous experts that span from clinicians to bioinformaticians, software developers, data miners and legal attorneys. All together we are defining specific scenarios to be used to test tools of the p-medicine infrastructure. SIB-BCF is defining the criteria and the guidelines to support and coordinate the evaluation process. An example of scenario: the dataset GSE22138 (http://www.ncbi.nlm.nih.gov/geo/) was downloaded and further analysed under several aspects.
It contains expression data from uveal melanoma primary tumours, 28 patients that developed metastases and 29 without metastases, with at least 3-year follow-up. After normalization and quality control, differentially expressed genes have been extracted and survival analysis has been performed.
This scenario has at least four different views according with the user's category:
Discussion and conclusion
Dr. John Mendelshon, director of the MD Anderson Cancer Center (Prof. De Pinho will become director starting from August 2011) and chairman of WIN listed the main next steps WIN has to take [14]. It is of interest to see that many of these steps are common with those of p-medicine, like the following operating procedures:
It is also important to realize that WIN Symposium's key notes, presentations and discussions shared several main points:
Schilsky [15] concluded the Symposium by listing what we still do not know and we should concentrate our efforts on:
As published by MacConaill and Garraway [20], the endless and fast progress in technology and cancer genome biology strongly interacts and fuels the transformation in oncology diagnosis, clinical trial design and treatment. This process needs to be supported by ethical and legal developments. From the organization point of view, clinicians, scientific leaders and all the research community must be ready to face a new phase of education to interpret and use the information that will be produced. Legal issue as well as ways to reimburse the new personalized therapies must be addressed to be ready to turn in reality the many promises developed and that will be discovered to improve patient's lives.
What emerges from the recent literature and the 3rd WIN symposium is that a minority of leaders are well informed of the issues they have to face accordingly with technological, legal and ethical aspects; the new process of re-organization, training and education will be a major challenge in the near future.
As p-medicine consortium, we are glad to be a group of 20 partners that cover all the aspects and backgrounds that are involved in building an infrastructure that will help personalized medicine to become daily clinical practice, as well as helping the subjects involved in train themselves in participating in this new way of treating patients.
Acknowledgments
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement N° 270089.
Competing Interests
The authors have declared that no competing interests exist.
References
1. Tursz T,
2. Volinia S,
3. Nicoloso MS,
4. Paik S,
5 van ‘t Veer LJ,
6. Farmer P,
7. Wistuba II,
8. Hood L,
9. Weston AD,
10 Auffray C,
11 Gorini A,
12 Gavaghan D,
13 Coveney PV,
14. Mendelsohn J 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care 6–8 July 2011, Paris, France, Opening Keynote Lecture
15. Schilsk RL 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care 6–8 July 2011, Paris, France, synthesis and perspectives
16. Burgess M 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care
17. Friend S 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care
18. Kurzrock R 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care
19. Pennington S 3rd WIN SYMPOSIUM in personalized cancer medicine, a significant step forward in patient care
20 Macconaill LE,