Genetic susceptibility in childhood acute leukaemias: a systematic review
Gisele D Brisson1, Liliane R Alves2 and Maria S Pombo-de-Oliveira1
1Paediatric Haematology–Oncology Programme, Research Centre, Instituto Nacional de Câncer, Rio de Janeiro, Brazil, 20231050
2Pharmacy Service, Multiprofessional Residency Programme, Instituto Nacional de Câncer, Rio de Janeiro, Brazil, 20231050
Correspondence to: Maria S. Pombo-de-Oliveira. Email: mpombo@inca.gov.br
Abstract
Acute leukaemias (AL) correspond to 25–35% of all cancer cases in children. The aetiology is still sheltered, although several factors are implicated in causality of AL subtypes. Childhood acute leukaemias are associated with genetic syndromes (5%) and ionising radiation as risk factors. Somatic genomic alterations occur during fetal life and are initiating events to childhood leukaemia. Genetic susceptibility has been explored as a risk factor, since environmental exposure of the child to xenobiotics, direct or indirectly, can contribute to the accumulation of somatic mutations. Hence, a systematic review was conducted in order to understand the association between gene polymorphisms and childhood leukaemia risk. The search was performed in the electronic databases PubMed, Lilacs, and Scielo, selecting articles published between 1995 and 2013. This review included 90 case-control publications, which were classified into four groups: xenobiotic system (n = 50), DNA repair (n = 16), regulatory genes (n = 15), and genome wide association studies (GWAS) (n = 9). We observed that the most frequently investigated genes were: NQO1, GSTM1, GSTT1, GSTP1, CYP1A1, NAT2, CYP2D6, CYP2E1, MDR1 (ABCB1), XRCC1, ARID5B, and IKZF1. The collected evidence suggests that genetic polymorphisms in CYP2E1, GSTM1, NQO1, NAT2, MDR1, and XRCC1 are capable of modulating leukaemia risk, mainly when associated with environmental exposures, such as domestic pesticides and insecticides, smoking, trihalomethanes, alcohol consumption, and x-rays. More recently, genome wide association studies identified significant associations between genetic polymorphisms in ARID5B e IKZF1 and acute lymphoblastic leukaemia, but only a few studies have replicated these results until now. In conclusion, genetic susceptibility contributes to the risk of childhood leukaemia through the effects of gene–gene and gene–environment interactions.
Keywords: leukaemia, genetic polymorphism, genetic predisposition to disease, environmental exposure
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 14/05/2015; Received: 09/03/2015
Introduction
Childhood acute leukaemia
Acute leukaemias (AL) are the highest incidence malignancy in children and adolescents (≤19 years of age) and as a whole, the aetiology has not yet been unveiled. There are two major groups of AL, acute lymphoblastic leukaemia (ALL) and acute myeloid leukaemia (AML), classified according to characteristics presented by leukaemic cells, such as morphological features, surface antigens, chromosomal and molecular abnormalities [1], and gene expression profile [2].
Observational epidemiology has demonstrated that about 5 to 10% of AL are associated with ionising radiation exposure and congenital genetic syndromes (Down, neurofibromatosis, Fanconi anaemia, and Bloom syndrome) [3], that are associated with specific leukaemia subtypes. For the remaining 90% of AL, the aetiopathology is postulated to be a multistep process and somatic mutations are the start point of the pathway. When initiated during fetal life, the majority of them require postnatal events that contribute to accumulation of secondary mutations and proliferative advantage [3–5]. In this regard, the initiating event originating in cells would take advantage of the genetic predisposition conferred, in part, by genetic susceptibility and damage from exogenous exposures [6].
Genetic susceptibility
Genetic polymorphisms are defined as natural genetic variations that occur randomly in the general population. The most common type is the single nucleotide polymorphism (SNP) that consists of a variation at a single base pair [7]. Depending on where it is located, SNPs can interfere with a gene’s function, affecting metabolic pathways [8]. This review will focus on three main pathways that have been related to AL genetic susceptibility: xenobiotic system, DNA repair system, and cell regulation, which have been identified as risk factors in childhood leukaemia.
Xenobiotic system
Children are more vulnerable and susceptible to environmental toxicants than adults because of physiological immaturity, and also indirect and unintended exposures. Environmental agents such as tobacco and traffic smoke, pesticides, household chemicals, paintings, and diet are potential AL risk factors, as they may contain carcinogenic substances to humans, such as organic solvents (benzene derivatives), polycyclic aromatic hydrocarbons (PAHs), and organochloride compounds [9]. These substances, however, require metabolic activation by enzymes from the xenobiotic system to be able to interact with genetic material and eventually cause somatic mutations [10].
The xenobiotic system is classified into two phases: i) phase I enzymes, represented by cytochrome P450 isoenzymes (CYP), that catalyse hydrolysis, reduction, and oxidation reactions; and ii) phase II enzymes, that catalyse conjugation reactions, comprising glutathione S-transferases (GST) and N-acetyl transferases (NAT).
Interindividual genetic variations that are capable of altering the process of metabolism of pro-carcinogens, in both mother and child, may modulate the risk of developing paediatric leukaemia [10].
DNA repair
DNA repair systems play an essential role in maintaining integrity and genomic stability [11]. Ionising radiation, environmental carcinogens and their reactive intermediates, together with genomic instability and inherent errors in DNA replication process contribute to the occurrence of damage in DNA. Mutations, chromosomal breaks, and crosslinks are actively recognised and repaired by sets of enzymes that constitute the DNA repair system [12]. Three main mechanisms are responsible for single-stranded DNA damage repair: i) base excision repair (BER), ii) nucleotide excision repair (NER), iii) and mismatch repair (MMR); which comprises enzymes enconded by several genes, such as XRCC1, ERCC2, MLH1, MSH3. Meanwhile, double-stranded DNA breaks can be repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ), throughout enzymes as nibrin (NBN) and others, encoded by several genes, such as ATM, BRCA2, and RAD51 [13]. Failures in those systems have been linked to birth defects, cancer, and premature ageing [12]. For instance, Fanconi anaemia, ataxia–telangiectasia, Nijmegen breakage syndrome, and Bloom syndrome, resulting from DNA repair disorders, are highly associated with childhood leukaemia [14].
Regulatory genes
Cells often present signal transduction alterations that lead to proliferation in response to external factors. Several growth factors, their receptors and effector molecules have been identified as proto-oncogenes or tumour suppressor genes. Mutations in these genes may interfere with regulatory mechanisms that control cell cycle, leading to generation of malignant clones [15]. Considering this fact, polymorphisms in genes involved in cell cycle regulation also contribute to cancer susceptibility [16]. Nevertheless, few studies to date have investigated the association of regulatory genes with paediatric leukaemia.
Considering the large amount of epidemiologic data about this subject, it becomes necessary to use systematic methods to evaluate and synthesise all the information in order to facilitate communication between molecular epidemiology and clinical practice [17]. In an attempt to clarify some issues in this field, a systematic review was conducted which aimed to add comprehensive information about genetic susceptibility in childhood leukaemia.
Methods
Publication search strategy
A literature search on genetic susceptibility and childhood leukaemia was carried out using PubMed, Lilacs, and Scielo (last updated in June 2013). The following terms were used in different combinations: ‘acute lymphoblastic leukaemia’, ‘acute myeloid leukaemia’, ‘genetic polymorphism(s)’, ‘genetic susceptibility’, ‘xenobiotic(s)’, ‘molecular epidemiology’, ‘risk factor(s)’, and ‘child* or infant or paediatric’. Also, the following MeSH terms were used: ‘precursor cell lymphoblastic leukaemia–lymphoma’, ‘leukaemia, myeloid, acute’, ‘polymorphism, genetic’, ‘genetic predisposition to disease’, ‘gene–environment interaction’, ‘case-control studies’, and ‘genetic association studies’. References were also checked in order to look for articles that were missing in the electronic databases. The search strategy was elaborated using the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement as a guideline [18].
Inclusion and exclusion criteria
The inclusion criteria for all publications were: 1) case-control genotyping studies published between 1995 and 2013 that tested the risk of genetic polymorphisms with childhood ALL and/or AML (ages ≤ 21 years); 2) studies that provided sufficient data for estimating the risk association with odds ratio (OR), relative risk (RR) or interaction odds ratio (IOR); and 3) full text available in English, Spanish, and Portuguese. The exclusion criteria were: 1) studies with a different theme from what was proposed for this review; 2) publications in different languages, otherwise the ones specified; 3) studies that specifically include leukaemia cases related to genetic syndromes—Down, neurofibromatosis, Fanconi anaemia, Bloom syndrome and ataxia–telangiectasia—or secondary leukaemia; 4) articles that include other malignancies besides leukaemia in the same cohort, avoiding extrapolation of results exclusively for leukaemias; 5) articles about family gene transmissions; 6) articles about genes and prognosis; 7) articles about folate genes and immune system; and 8) comments and editorials.
Data extraction
Information was extracted from each eligible article supervised by two investigators (LRA and MSPO), according to the inclusion criteria listed above. The following variables were collected from eligible studies: geographical origin, first author’s name, year of publication, leukaemia subtype, number of cases and controls, age, candidate genes investigated, and significant genotyping results.
Statistical analysis
The strength of association between different genetic polymorphisms through the case-control method was evaluated by analysis of OR, RR, or IOR, with 95% confidence interval (95% CI), that were collected from the studies. Risk associations were considered significant when the P-value was ≤0.05.
Results and discussion
After screening of the retrieved titles, 312 publications were identified as potentially relevant to this review (Figure 1). After application of exclusion criteria, 103 publications were pre-selected, of which 22 were reviews, and 81 were case-control studies. After checking for bibliographies of pre-selected publications, nine papers were added, giving a total of 90 case-control publications included in this review. The majority of publications addressed polymorphisms in genes related to xenobiotic system (n = 50), followed by DNA repair genes (n = 16), regulatory genes (n = 15), and genome wide association studies (GWAS) (n = 9). The most frequently analysed gene polymorphisms, presented by at least three papers (Figure 2), were located in genes CYP1A1, CYP2D6, CYP2E1, CYP3A5, EPHX1, GSTM1, GSTT1, GSTP1, MPO, NAT2, NQO1, MDR1 (ABCB1), XRCC1, ERCC2, NBN, ARID5B, and IKZF1. Figure 2 also shows the proportion of publications that showed statistically significant associations for each gene, among the total. Considering only statistically significant results, median ORs for increased risk or protective associations for those genes were calculated, and are demonstrated in Figure 3.
Xenobiotic system
Genetic susceptibility studies related to xenobiotic system are presented in Table 1. The main investigated gene polymorphisms comprised the genes CYP1A1, CYP2D6, CYP2E1, CYP3A4, CYP3A5, EPHX1, GSTM1, GSTP1, GSTT1, MDR1, MPO, NAT1, NAT2, and NQO1. Most publications are from Asia (39.6%), followed by North America (25%), Europe (20.8%), and South America (14.6%). The great disparity between ethnic groups is remarkable, since the Asian continent includes a wide range of people with distinct genetic backgrounds, such as Caucasians, Turkish, Indians, Japanese, Chinese, and the Korean population, likewise, Americans have diverse ancestries, mainly Caucasian, Hispanic, and African. The vast majority of publications (75%) investigated the genetic susceptibility in ALL only, and 25% in AML ALL; no one has investigated AML solely.
Regarding phase I metabolism, fourteen publications [19–32] explored polymorphisms of CYP1A1, mainly the variant alleles *2A, *2B, *2C, and *4; only six (42.9%) found significant associations between these alleles and childhood ALL. CYP1A1*2A allele was associated with increased risk for ALL among Canadians [27], Indians [26], and Hispanics in North-America [31]. CYP1A1*2B was associated with increased risk for B-cell precursor ALL (Bcp-ALL) among north-Americans [31]. CYP1A1*2C allele was also associated with increased risk for ALL among Indians [26] and north-Americans [31]. Two Brazilian papers found significant associations of CYP1A1 variant alleles and ALL only with combined genotypes: CYP1A1*2 CYP2E1*5B GSTP1*B GSTM1-null [23], and CYP1A1*2A/*2B/*2C NQO1 609-CT/ CT TT [32].
Six publications [19, 22–24, 33, 34] explored variant alleles of CYP2E1, mainly *5B, *6 and *7B; in four out of six (66.7%), significant associations were found: CYP2E1*5B was related to increased risk for ALL/AML in Canadians [33] and Turkish [19]. The presence of at least two variant alleles (*5B and *6; *6 and *7B; or *5B, *6, and *7B) was related to increased risk for ALL in Turkish [34]. Besides, the combined genotype CYP2E1*5B CYP1A1*2 GSTP1*B GSTM1-null was associated with increased risk for ALL in Brazilians [23].
Six publications [19, 24–27, 35] explored CYP2D6*3 and *4 alleles. The wild-type allele (CYP2D6*1) was negatively associated with ALL in Brazilians [35]. Polymorphisms of CYP3A genes (CYP3A4*1B, CYP3A5*3, and CYP3A5*6) were investigated in four papers [24, 29, 36, 37], and no significant associations were found for CYP3A4*1B allele [24, 29]. The wild-type allele CYP3A5*1 was associated with ALL increased risk in Denmark and Norway [36], whereas alleles CYP3A5*3 and CYP3A5*6 were associated with ALL protection in Brazilians [37]. The same Brazilian study found increased risk association for the CYP3A5*6 allele only in Whites, leading to speculation that CYP3A5 may also be involved in detoxification as well as activation mechanisms of carcinogens. Alleles that do not produce a functional protein, such as CYP3A5*6, would contribute to accumulation of potentially harmful substances [36].
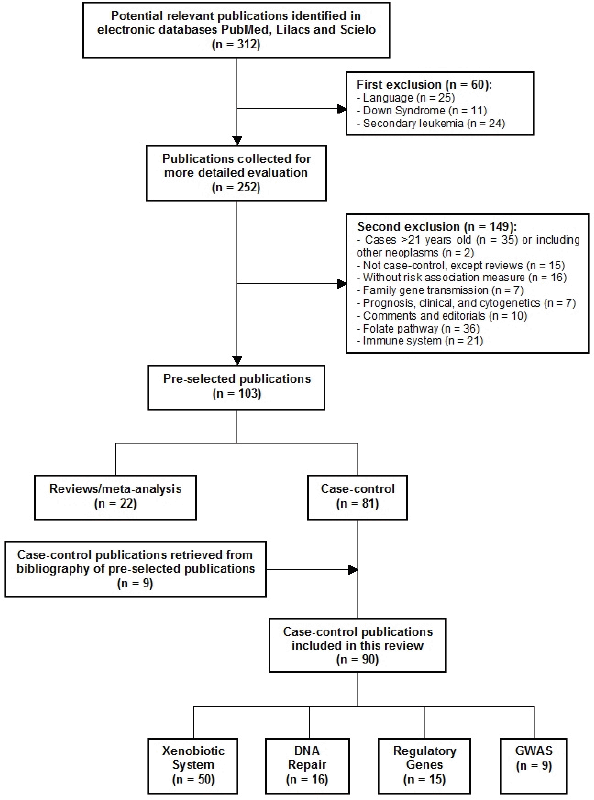
Figure 1. Flow diagram of included and excluded publications.
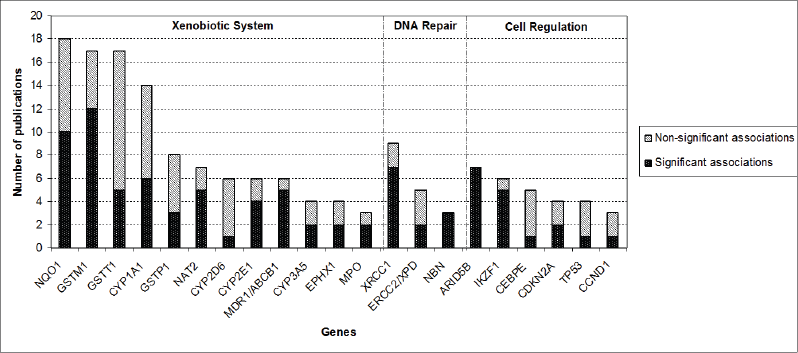
Figure 2. Proportion of publications that reported statistically significant associations between genetic polymorphisms and CAL risk for each of the genes that had polymorphism analysis reported by at least three publications.
Eighteen publications [21, 22, 24, 25, 32, 33, 35, 38–48] addressed polymorphisms of NQO1 gene (C609T and C465T). In ten studies (55.5%), divergent associations were found. NQO1 609T allele was associated with increased risk for ALL among Canadians [33] and Brazilians [32]. In Filipinos, the genotype NQO1 609CC was associated with increased risk for ALL [44], whereas the variant allele NQO1 609T was negatively associated with ALL in Malaysian boys [48] and Brazilians [35]. The 609T allele has been associated with infant leukaemia and MLL gene rearrangements in Caucasians [39, 42, 46, 47]. Otherwise, very few publications demonstrated increased risk for ALL in Canadians [33], and Japanese [40] with NQO1 465T allele.
Few publications [22, 24, 35, 49] have explored polymorphisms of EPHX1*2 (T28C–Tyr113His) and EPHX1*3 (A52G–His139Arg); two out of four showed divergent results: variant alleles *2 and *3 were associated with protection for ALL in Brazilians [35], whereas the *2 allele was associated with increased risk for ALL in Turkish [49]. Given the duality of functions that enzyme EPHX1 performs, its interconnection with CYP450 family, the diversity of xenobiotics presented in the environment, and the differences in allele frequencies among populations, gene polymorphisms of EPHX1 may contribute in an unpredictable way in activation or detoxification of xenobiotics.
Finally, three publications [24, 33, 35] demonstrated that the variant alleles of MPO, mainly MPO*2 (G-463A) have a protective effect when combined to other gene polymorphisms [33, 35]. No study has ever demonstrated an independent effect of MPO*2 in childhood leukaemia susceptibility.
Regarding phase II metabolism, the GST gene family was the most investigated (20 case-control publications), regarding association with childhood leukaemia risk all over the world. In twelve out of seventeen [19, 20, 23–27, 29, 38, 44, 48, 50–55] the homozygous deletion of GSTM1 and GSTT1 alleles (null genotype) was related to childhood leukaemia risk: GSTM1-null genotype was associated with increased risk for ALL in ten papers [23, 26, 27, 29, 38, 44, 50, 52, 55]; GSTT1-null genotype was associated with increased risk for ALL, mostly when combined with GSTM1-null genotype [29, 38, 52, 55]. However, two studies performed in US children disclosed conflicting results: GSTM1-null was found to be associated with protection for ALL among non-Hispanic American children [24], whereas non-null alleles GSTM1*A, GSTM1*B and GSTT1*1 were associated with increased risk for ALL [51]. These discrepancies may be because of differences in allele frequencies in mixed populations and the different patterns of environmental exposures.
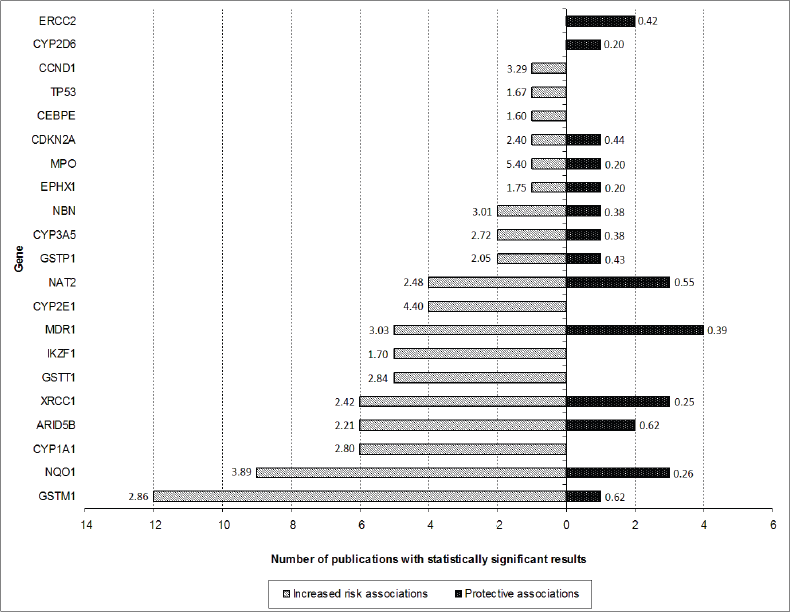
Figure 3. Number of publications that have shown statistically significant protective (black bar) or increased risk (white bar) associations for each gene (only genes with polymorphism analysis reported by at least three publications are shown). The numbers presented in the extremities of each bar represent the median ORs of risk associations found for polymorphisms involving each gene.
From eight publications [20, 23, 24, 38, 51, 54, 56, 57] that explored GSTP1 polymorphisms (A1578G–GSTP1*B; C2293T–GSTP1*C), only three (37.5%) showed significant associations, but with opposite results: GSTP1*B was related to increased risk for ALL among Canadians [57] and Brazilians [23], while a protective effect was observed for Indonesian girls [38]. Seven publications [22, 24, 25, 37, 58–60] analysed the complexity of NAT2 polymorphisms. Overall, haplotypes that result in low activity phenotype were associated with increased risk for childhood leukaemia [59]. The only exception was a protective effect of NAT2 341C-481T-590A, that results in slow activity phenotype, among Russians [25]. Besides, the combination of slow phenotype in both child and mother intensified the risk for early age ALL [60]. The rapid allele *4 was associated with protection for ALL among Canadians [58], as well as for AML in Brazilian children [59].
Table 1. Genetic susceptibility publications in childhood leukaemia involving genes related to xenobiotic system.
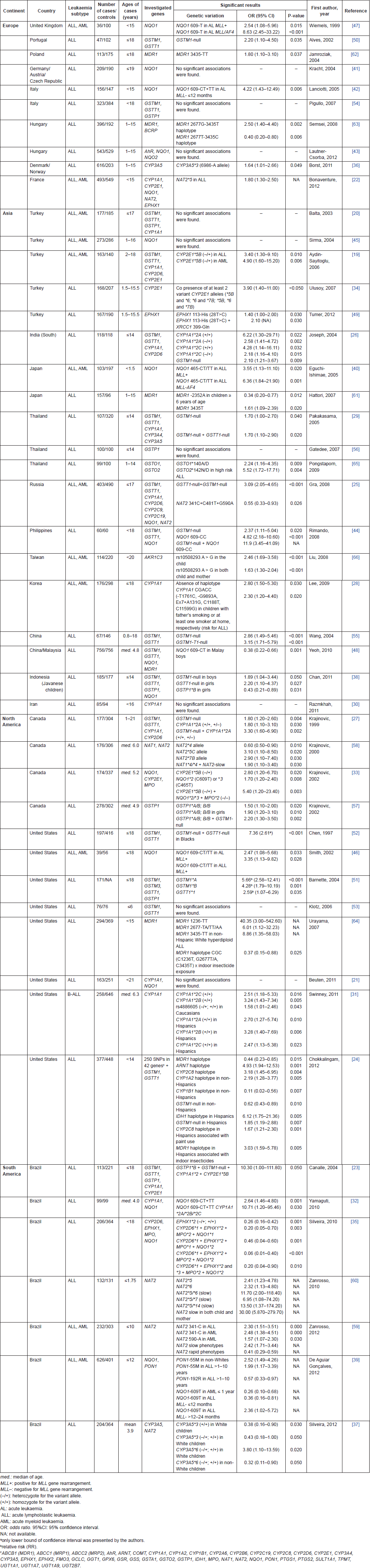
Besides the detoxifying function of some metabolising enzymes, membrane transporter proteins also act protectively against carcinogens. Six publications [24, 48, 61–64] explored polymorphisms of MDR1 (ABCB1 family) gene, mainly C1236T, G2677T/A, C3435T, and T-129C, that encodes an efflux membrane transporter (P-glycoprotein) with childhood ALL. MDR1 3435T allele was consensually associated with increased risk for ALL in four publications [61–64], while the haplotype GAGT (rs2520464, rs12334183, rs1202179, rs17327442) was associated with protection for ALL in north-Americans [24].
Finally, few publications have explored other genes, such as ARNT, CYP1A2, CYP1B1, CYP2C8 and IDH1 [24], PON1 [39], NAT1 [58], GSTO1, and GSTO2 [65], and AKR1C3 [66], in childhood ALL. Their results, however, need to be replicated in further studies.
DNA repair
The data from genotyping studies in genes related to DNA repair system are summarised in Table 2. The main investigated gene polymorphisms comprised the genes ERCC2, MLH1, MSH3, NBN, and XRCC1. Six publications (37.5%) were performed in Europeans, five (31.25%) in Asians, four (25%) in north-Americans, and one (6.25%) in Brazilians. The majority of them focused only in ALL.
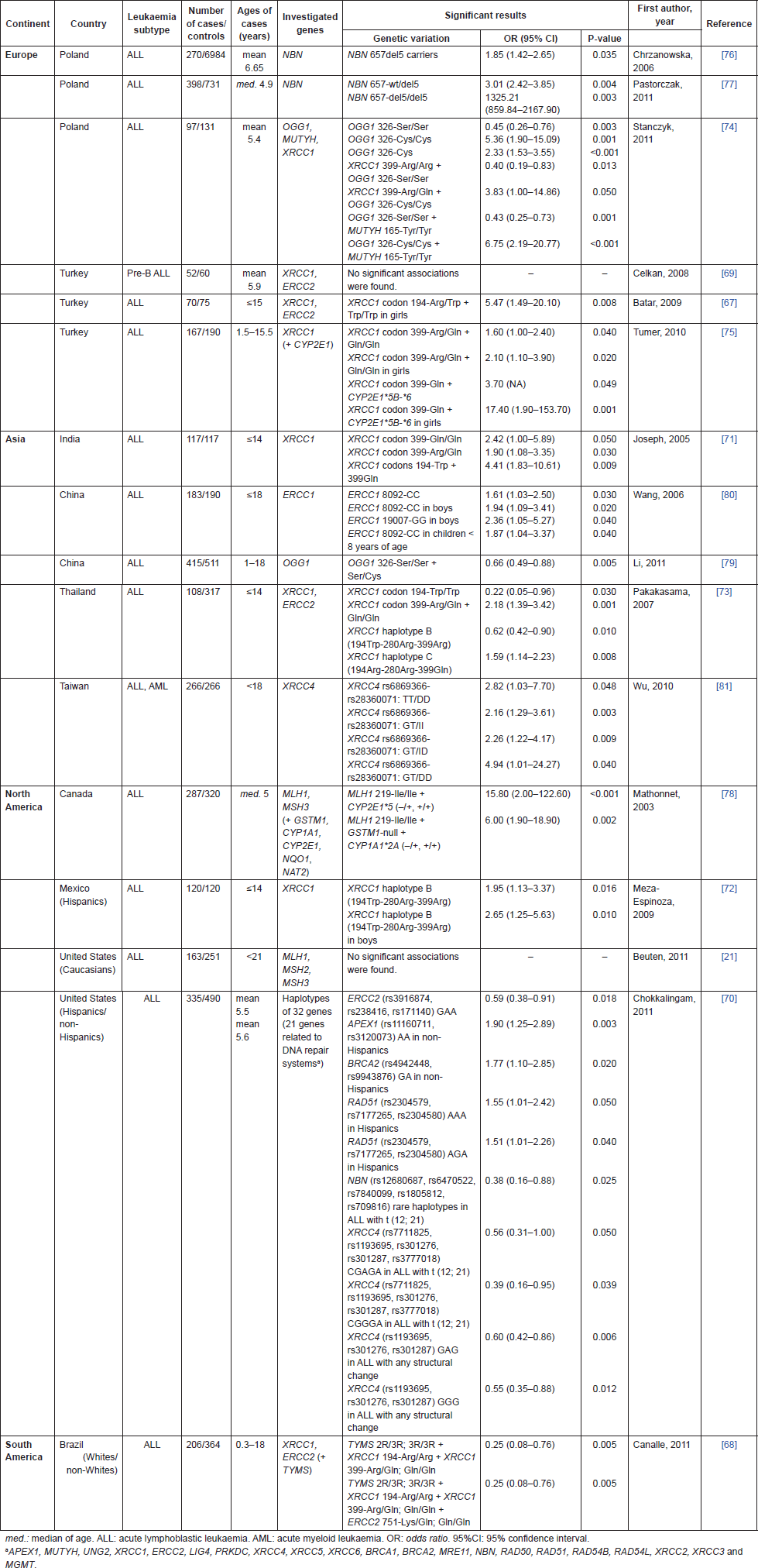
Nine case-control publications [67–75] addressed polymorphisms of XRCC1 (Arg194Trp, Arg280His, and Arg399Gln), which encodes a protein involved in BER pathway. Seven of them (77.8%) have shown significant associations with childhood ALL. XRCC1 194Trp allele was related to increased risk among Turkish girls [67], Indians [71], and Mexicans [72], while it was related to protection among Thai [73]. No publication showed association of 280His allele with childhood ALL. XRCC1 399Gln allele was related to increased risk for ALL among Indians [71], Thai [73], Turkish [75], and Poles [74]. However, when combined with XRCC1 194Arg wild-type allele and the variant alleles ERCC2 751Gln and TYMS 3R, the XRCC1 399Gln allele was related to protection for ALL among Brazilians [68].
The ERCC2 gene encodes a DNA helicase involved in NER pathway, and its polymorphisms (Asp312Asn and Lys751Gln), were explored in five case-control publications [67–70, 73]. No one was able to show an independent association with leukaemia risk. ERCC2 751Gln allele was related to protection for ALL among Brazilians when combined with XRCC1 399Gln-194Trp and TYMS 3R [68]. The haplotype GAA (rs3916874, rs238416, rs171140) was also associated with protection for ALL among north-Americans [70].
Genetic polymorphisms of NBN, which is involved in DNA repair by HR, were reported by three publications [70, 76, 77]. Two of them showed that 657del5 mutation was related to increased risk for ALL among Poles [76, 77]. Recently, five SNPs of NBN gene (rs12680687, rs6470522, rs7840099, rs1805812, rs709816) were associated with protection for a subset of Bcp-ALL in north-Americans [70], reinforcing that the interaction of multiple polymorphisms can influence paediatric leukaemia risk.
Polymorphisms of two genes involved in MMR, MLH1, and MSH3, were explored in two papers [21, 78], but both failed to demonstrate any independent association with childhood ALL. However, the combination of MLH1 219-Ile/Ile with genetic variants of CYP2E1 or GSTM1 and CYP1A1 increased the risk for ALL [78]. Other genotyping studies of OGG1 [74, 79], MUTYH [74], ERCC1 [80], XRCC4 [70, 81], APEX1, BRCA2, and RAD51 [70] found increased associations with childhood ALL.
Regulatory genes and GWAS
To date, the majority of publications regarding regulatory genes and childhood leukaemia were performed to validate GWAS results. The main investigated gene polymorphisms are summarised in Table 3. The majority of publications focused only in ALL (87.5%) and is from North America, followed by Asians (25%), and Europeans (25%).
Prior to GWAS publications, a few genetic polymorphisms of regulatory genes were investigated by candidate gene approach. CCND1 870AA genotype (homozygous wild-type) was related to increased risk for ALL among Chinese [82]. Polymorphisms in promoter regions of genes CDKN2A (T-222A), CDKN2B (C-1270T, A-593T, C-287G) and CDKN1B (G-1608A) were also associated with childhood pre-B ALL risk among Canadians [16]. Furthermore, the Arg72Pro polymorphism in TP53 gene was associated with increased risk for ALL among children from the United Kingdom [83]. Polymorphisms of other genes were also related to childhood acute leukaemia (CAL) risk, like BAT3 and DAXX [83], ATM [84], TERT [85], MDM2 [86], LMO1 [21], MLL and EP300 [87], but the associations need to be replicated in further studies.
Table 2. Genetic susceptibility publications in childhood leukaemia involving genes related to DNA repair.
The first two GWAS regarding genetic susceptibility to childhood leukaemia were published in 2009, which observed that SNPs in regions 7p12.2 (IKZF1 rs4132601, rs11978267), 10q21.2 (ARID5B rs7089424, rs10821936, rs10994982), and 14q11.2 (CEBPE rs2239633) were associated with childhood ALL risk, specifically with B-cell acute lymphoblastic leukaemia (B-ALL), and hyperdiploid subsets, with ORs ranging from 1.34 to 1.91, and P <10–7 [88, 89]. In subsequent analysis, the association of CDKN2A rs3731217 (T allele) with protection for ALL (OR 0.71, P = 3.01 x 10–11) was validated [90], and ARID5B rs10821936 was associated with ALL among Blacks (OR 2.08, P = 0.0015), mainly hyperdiploid B-ALL (OR 6.62, P <0.001) in the US [91]. Also, it was demonstrated that 24% of the total variation in B-cell precursor acute lymphoblastic leukaemia (Bcp-ALL) risk is accounted for common genetic variation, which supports for a polygenic basis for susceptibility to Bcp-ALL [92]. French studies have shown similar associations regarding IKZF1 and ARID5B, and also found associations between CDKN2A rs3731217 (OR 0.8) and CEBPE rs2239633 (OR 0.9) with ALL risk [93].
Since then, other research groups have aimed to replicate the risk associations previously identified by the GWAS. As shown in Table 3, seven papers confirmed ARID5B rs10821936 association with increased ALL risk, mainly B-ALL [43, 77, 94–98]. From six publications that explored IKZF1 SNPs [21, 43, 77, 95–97], five of them confirmed the risk associations for ALL. CDKN2A and CEBPE SNPs associations were replicated by one study respectively [70, 95].
SNPs in other genes were also identified as risk variants for ALL by other GWAS, but replication of these results are still needed: HAO1 (rs6140264, OR 8.84), EPB41L2 (rs9388856, rs9388857, rs1360756, OR 8.97), C2orf3 (rs12105972, OR 0.13), and MAN2A1 (rs3776932, OR 0.11) were associated with ALL risk among Koreans (P = 0.0001) [99]; TP63 (rs17505102, OR 0.63, P = 4.87 x 10–7), PTPRJ (rs3942852, OR 0.77, P = 2.54 x 10–4), and EPOR (rs4804164, OR 0.58, P = 0.008; rs317913, OR 0.60, P = 0.019) were associated with ETV6/RUNX1 ALL risk among Europeans [100, 101].
Gene-environment interaction
Few publications addressed the interaction between gene polymorphisms, environmental exposures, and childhood leukaemia. Studies performed in children exposed directly and/or indirectly throughout maternal exposures are scarce. To date, environmental exposures that have been explored were smoking (tobacco exposure), pesticides, insecticides, trihalomethanes (chlorination by-products of drinking water) alcohol consumption, paint use and x-rays. The absence of CYP1A1 CGACC haplotype, consisting of five SNPs (-T1761C, -G9893A, Ex7 A131G, C1188T, C11599G), was associated with increased risk for ALL among children with father’s smoking or at least one smoker at home [28]. Also, haplotypes of CYP2C8 and MDR1 (ABCB1) were related to increased risk for ALL when associated with paint use and indoor insecticides, respectively among Hispanics living in the US [24]. Another finding was that children exposed to indoor insecticides carrying MDR1 haplotype CGC (C1236T, G2677T/A, C3435T) presented a lower risk for ALL [64], indicating that pesticides’ toxic effects may be influenced by efflux through P-glycoprotein complex.
Six publications [102–107] estimated risk associations by calculating IOR in a case-only cohort. It was observed that CYP1A1*2A/*2B increased the risk for ALL by five-fold among children exposed to pesticides during maternal pregnancy and childhood, while CYP1A1*2B was related to a protective effect among children with mothers who had smoked 1–20 cigarettes during the first trimester of pregnancy (IOR = 0.1; IC95% 0.01–0.9) and with fathers who had smoked >20 cigarettes between birth and diagnosis (IOR 0.2; IC95% 0.04–0.9) [102, 103]. Also, it was observed that among children exposed to higher levels of trihalomethanes in drinking water, the risk for ALL was increased in the presence of the polymorphic variant CYP2E1*5 (IOR 9.75; IC95% 1.10–86.01), and GSTT1 deletion (IOR 9.13; IC95% 1.44–57.82), in pre and post-natal periods, respectively [104]. GSTM1-null genotype and CYP2E1*5 variant were related to increased risk for ALL among children with mothers who had consumed alcoholic beverages during the third trimester of pregnancy (IOR 2.4; IC95% 1.1–5.4) and nursing period (IOR 4.9; IC95% 1.4–16.6), respectively [105]. Thus, it is notable that variations in xenobiotic metabolism resulted from genetic polymorphisms can modulate childhood leukaemia risk.
Concerning DNA repair genes, it was observed that variants of APEX1 (Asp148Glu) and MLH1 (Ile219Val) were associated with a protective effect for ALL among girls exposed to x-rays (one or more exposures) during postnatal period (MLH1 IOR 0.2; IC95% 0.1–0.8; and APEX1 IOR 0.1; IC95% 0.0–0.7) [106, 107]. Also, the protective effect for ALL of XRCC4 GGG haplotype, consisting of SNPs rs1193695, rs301276, and rs301287, was modulated by number of postnatal x-rays (P = 0.027) [70]. Again, cancer susceptibility resulted from the interaction of environmental exposure and genetic polymorphism, which highlights the multifactorial aetiology of paediatric leukaemia.
Conclusions
Great scientific advances in the understanding of paediatric leukaemia have been made. Unlike the adult, who usually develops cancer because of the cumulative effect of environmental exposures during his life, the child, which manifests leukaemia with a short latency period, does not have enough exposure time to allow the initiation of a long carcinogenic process. Thus, genetic susceptibility may play an important role in modulating environmental exposures’ effects.
This systematic review gathered publications up to 2013 and was an attempt to overview the risk associations between several gene polymorphisms and paediatric AL. It was possible to collect from the selected studies significant amount of data, which is considered to be a fair representation of international scientific literature on this subject. The vast majority of studies so far focused on evaluating the magnitude of risk of genetic polymorphisms in ALL, mainly because Bcp-ALL is the most frequent type of leukaemia in children. In this context, we also realised that there is still a great need for further investigations on the risk factors for paediatric AML.
Regarding the xenobiotic system, gene polymorphisms of CYP2E1, GSTM1, NQO1, NAT2, and ABCB1 (MDR1) were more frequently associated with childhood leukaemia risk, which also showed interaction effect with environmental exposures such as paints, household pesticides, insecticides, smoking, alcohol, and trihalomethanes. Gene polymorphisms related to DNA repair have been little investigated in paediatric leukaemia, maybe because of its association with genetic diseases. However, it was noticed that XRCC1 polymorphisms play an important role in the development of ALL, and postnatal exposure to x-rays can modulate leukaemia risk in the presence of APEX1, MLH1, and XRCC4 gene variants. While interpreting these results, one has to consider that fetuses and infants are naturally more affected than adults by a variety of environmental toxicants, mainly because of differential exposure and physiologic immaturity, which makes them more susceptible to suffer from DNA damage and less capable of detoxifying carcinogenic compounds [108]. And so, genetic polymorphisms involving xenobiotic and DNA repair systems have a major role in modulating the effects of environmental agents in children.
Some limitations were observed in the studies that might make the consolidation of scientific evidence difficult, such as: 1) relatively small number of cases, making it difficult to obtain statistically significant results; 2) ethnic and racial differences between populations, which are reflected in distinct polymorphic allele frequencies and patterns of exposure to environmental agents; 3) little information regarding the effect of gene polymorphisms on the encoded protein; 4) and few considerations about gene–environment interactions. Also, we could observe that the majority of associations provided low risk estimates (OR <2.0), which showed that gene polymorphisms are of low penetrance, and conceptually, are minor parts of multifactorial pathways to childhood leukaemia [6].
More recently, GWAS have identified new gene polymorphisms potentially related to paediatric ALL, particularly involving IKZF1 and ARID5B, which were subsequently replicated in independent studies. As a result, we see genetic susceptibility clearly contributes to childhood leukaemia risk, mainly through gene–gene and gene–environment interactions. Further studies are still needed to confirm the observed associations in different populations and to characterise environmental agents as risk factors for childhood leukaemia.
Conflicts of interest
The authors declare no conflict of interest.
Authors’ contributions
The study was designed by GDB, LRA, and MSPO. The literature search and data analysis were carried out by GDB, with the supervision of LRA and MSPO. The manuscript was prepared by GDB, LRA, and MSPO.
References
1. Greaves M (2002) Childhood leukaemia BMJ 324(7332) 283–7 DOI: 10.1136/bmj.324.7332.283 PMID: 11823363 PMCID: 1122200
2. Mullighan CG, Flotho C and Downing JR (2005) Genomic assessment of pediatric acute leukemia Cancer J 11(4) 268–82 DOI: 10.1097/00130404-200507000-00003 PMID: 16197717
3. Wiemels J (2012) Perspectives on the causes of childhood leukemia Chem Biol Interact 196(3) 59–67 DOI: 10.1016/j.cbi.2012.01.007 PMID: 22326931 PMCID: 3839796
4. Greaves M (1999) Molecular genetics, natural history and the demise of childhood leukaemia Eur J Cancer 35(14) 1941–53 DOI: 10.1016/S0959-8049(99)00296-8
5. Greaves M (2005) In utero origins of childhood leukaemia Early Hum Dev 81(1) 123–9 DOI: 10.1016/j.earlhumdev.2004.10.004 PMID: 15707724
6. Chokkalingam AP and Buffler PA (2008) Genetic susceptibility to childhood leukaemia Radiation protection dosimetry 132(2) 119–29 DOI: 10.1093/rpd/ncn255 PMID: 18922824 PMCID: 2879095
7. Polymorphism [http://ghr.nlm.nih.gov/glossary=polymorphism] date accessed 14 August 2014
8. What are single nucleotide polymorphisms (SNPs)? [http://ghr.nlm.nih.gov/handbook/genomicresearch/snp] date accessed: 14 August 2014
9. Belson M, Kingsley B and Holmes A (2007) Risk factors for acute leukemia in children: a review Environ Health Perspect 115(1) 138–45 DOI: 10.1289/ehp.9023 PMID: 17366834 PMCID: 1817663
10. Sinnett D, Krajinovic M and Labuda D (2000) Genetic susceptibility to childhood acute lymphoblastic leukemia Leuk Lymphoma 38(5–6) 447–62 DOI: 10.3109/10428190009059264 PMID: 10953966
11. Wang L et al (2012) X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and risk of childhood acute lymphoblastic leukemia: a meta-analysis PloS one 7(4) e34897 DOI: 10.1371/journal.pone.0034897 PMID: 22529951 PMCID: 3329555
12. Ronen A and Glickman BW (2001) Human DNA repair genes Environ Mol Mutagen 37(3) 241–83 DOI: 10.1002/em.1033 PMID: 11317342
13. Morita R et al (2010) Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems J Nucleic Acids 2010 179594 DOI: 10.4061/2010/179594 PMID: 20981145 PMCID: 2957137
14. Seif AE (2011) Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis Cancer Genet 204(5) 227–44 DOI: 10.1016/j.cancergen.2011.04.005 PMID: 21665176
15. Malumbres M and Barbacid M (2001) To cycle or not to cycle: a critical decision in cancer Nat Rev Cancer 1(3) 222–31 DOI: 10.1038/35106065
16. Healy J et al (2007) Promoter SNPs in G1/S checkpoint regulators and their impact on the susceptibility to childhood leukemia Blood 109(2) 683–92 DOI: 10.1182/blood-2006-02-003236
17. Khoury MJ et al (2013) Transforming epidemiology for 21st century medicine and public health Cancer Epidemiol Biomarkers Prev 22(4) 508–16 DOI: 10.1158/1055-9965.EPI-13-0146 PMID: 23462917 PMCID: 3625652
18. Moher D et al (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement BMJ 339 b2535 DOI: 10.1136/bmj.b2535 PMID: 19622551 PMCID: 2714657
19. Aydin-Sayitoglu M et al (2006) Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes in the susceptibility to acute leukemias Am J Hematol 81(3) 162–70 DOI: 10.1002/ajh.20434 PMID: 16493615
20. Balta G et al (2003) Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia Am J Hematol 73(3)154–60 DOI: 10.1002/ajh.10339 PMID: 12827651
21. Beuten J et al (2011) Candidate gene association analysis of acute lymphoblastic leukemia identifies new susceptibility locus at 11p15 (LMO1) Carcinogenesis 32(9)1349–53 DOI: 10.1093/carcin/bgr091 PMID: 21602560
22. Bonaventure A et al (2012) Maternal smoking during pregnancy, genetic polymorphisms of metabolic enzymes, and childhood acute leukemia: the ESCALE study (SFCE) Cancer Causes Control 23(2) 329–45 DOI: 10.1007/s10552-011-9882-9
23. Canalle R et al (2004) Genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia Environ Mol Mutagen 43(2) 100–9 DOI: 10.1002/em.20003 PMID: 14991750
24. Chokkalingam AP et al (2012) Variation in xenobiotic transport and metabolism genes, household chemical exposures, and risk of childhood acute lymphoblastic leukemia Cancer Causes Control 23(8) 1367–75 DOI: 10.1007/s10552-012-9947-4 PMID: 22674224 PMCID: 3390694
25. Gra OA et al (2008) [Genetic polymorphism in GST, NAT2, and MTRR and susceptibility to childhood acute leukemia] Molekuliarnaia biologiia 42(2) 214–25 PMID: 18610829
26. Joseph T et al (2004) Genetic polymorphism of CYP1A1, CYP2D6, GSTM1 and GSTT1 and susceptibility to acute lymphoblastic leukaemia in Indian children Pediatr Blood Cancer 43(5) 560–7 DOI: 10.1002/pbc.20074 PMID: 15382273
27. Krajinovic M et al (1999) Susceptibility to childhood acute lymphoblastic leukemia: influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 genetic polymorphisms Blood 93(5) 1496–501 PMID: 10029576
28. Lee KM et al (2009) Paternal smoking, genetic polymorphisms in CYP1A1 and childhood leukemia risk Leuk Res 33(2) 250–8 DOI: 10.1016/j.leukres.2008.06.031 PMCID: 2787091
29. Pakakasama S et al (2005) Polymorphisms of drug-metabolizing enzymes and risk of childhood acute lymphoblastic leukemia Am J Hematol 79(3) 202–5 DOI: 10.1002/ajh.20404 PMID: 15981231
30. Razmkhah F et al (2011) Frequency of CYP1A1*2C Polymorphism in Patients with Leukemia in the Iranian Population Lab Med 42 220–3 DOI: 10.1309/LM337JWOSVNEHPUI
31. Swinney RM et al (2011) Polymorphisms in CYP1A1 and ethnic-specific susceptibility to acute lymphoblastic leukemia in children Cancer Epidemiol Biomarkers Prev 20(7) 1537–42 DOI: 10.1158/1055-9965.EPI-10-1265 PMID: 21586621
32. Yamaguti GG et al (2010) Increased risk for acute lymphoblastic leukemia in children with cytochrome P450A1 (CYP1A1)- and NAD(P)H:quinone oxidoreductase 1 (NQO1)-inherited gene variants Acta Haematol 124(3) 182–4 DOI: 10.1159/000320275 PMID: 20962519
33. Krajinovic M et al (2002) Role of NQO1, MPO and CYP2E1 genetic polymorphisms in the susceptibility to childhood acute lymphoblastic leukemia Int J Cancer 97(2) 230–6 DOI: 10.1002/ijc.1589 PMID: 11774269
34. Ulusoy G et al (2007) Significance of genetic polymorphisms at multiple loci of CYP2E1 in the risk of development of childhood acute lymphoblastic leukemia Oncology 72(1–2) 125–31 DOI: 10.1159/000111131
35. Silveira Vda S et al (2010) Role of the CYP2D6, EPHX1, MPO, and NQO1 genes in the susceptibility to acute lymphoblastic leukemia in Brazilian children Environ Mol Mutagen 51 48–56
36. Borst L et al (2011) The impact of CYP3A5*3 on risk and prognosis in childhood acute lymphoblastic leukemia Eur J Haematol 86(6) 477–83 DOI: 10.1111/j.1600-0609.2011.01608.x PMID: 21418106
37. Silveira VS et al (2012) CYP3A5 and NAT2 gene polymorphisms: role in childhood acute lymphoblastic leukemia risk and treatment outcome Mol Cell Biochem 364(1–2) 217–23 DOI: 10.1007/s11010-011-1220-8 PMID: 22215203
38. Chan JY et al (2011) Xenobiotic and folate pathway gene polymorphisms and risk of childhood acute lymphoblastic leukaemia in Javanese children Hematol Oncol 29(3) 116–23 DOI: 10.1002/hon.965
39. De Aguiar Goncalves BA et al (2012) NQO1 rs1800566 (C609T), PON1 rs662 (Q192R), and PON1 rs854560 (L55M) polymorphisms segregate the risk of childhood acute leukemias according to age range distribution Cancer Causes Control 23(11) 1811–9 DOI: 10.1007/s10552-012-0060-5 PMID: 22976839
40. Eguchi-Ishimae M et al (2005) The association of a distinctive allele of NAD(P)H:quinone oxidoreductase with pediatric acute lymphoblastic leukemias with MLL fusion genes in Japan Haematologica 90(11) 1511–5 PMID: 16266898
41. Kracht T et al (2004) NQO1 C609T polymorphism in distinct entities of pediatric hematologic neoplasms Haematologica 89(12) 1492–7 PMID: 15590400
42. Lanciotti M et al (2005) Genetic polymorphism of NAD(P)H:quinone oxidoreductase is associated with an increased risk of infant acute lymphoblastic leukemia without MLL gene rearrangements Leukemia 19(2) 214–6 DOI: 10.1038/sj.leu.2403613
43. Lautner-Csorba O et al (2012) Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance BMC Med Genomics 5 42 DOI: 10.1186/1755-8794-5-42 PMID: 23021489 PMCID: 3542204
44. Rimando MG et al (2008) Prevalence of GSTT1, GSTM1 and NQO1 (609C > T) in Filipino children with ALL (acute lymphoblastic leukaemia) Biosci Rep 28 117–24 DOI: 10.1042/BSR20070010 PMID: 18444911
45. Sirma S, Agaoglu L, Yildiz I, Cayli D, Horgusluoglu E and Anak S et al (2004) NAD(P)H:quinone oxidoreductase 1 null genotype is not associated with pediatric de novo acute leukemia Pediatr Blood Cancer 43(5) 568–70 DOI: 10.1002/pbc.20098 PMID: 15382274
46. Smith MT et al (2002) Low NAD(P)H:quinone oxidoreductase activity is associated with increased risk of leukemia with MLL translocations in infants and children Blood 100(13) 4590–3 DOI: 10.1182/blood-2001-12-0264 PMID: 12393620
47. Wiemels JL et al (1999) A lack of a functional NAD(P)H:quinone oxidoreductase allele is selectively associated with pediatric leukemias that have MLL fusions Cancer Res 59(16) 4095–9 PMID: 10463613
48. Yeoh AE et al (2010) Genetic susceptibility to childhood acute lymphoblastic leukemia shows protection in Malay boys: results from the Malaysia-Singapore ALL Study Group Leuk Res 34(3) 276–83 DOI: 10.1016/j.leukres.2009.07.003
49. Tumer TB, Sahin G and Arinc E (2012) Association between polymorphisms of EPHX1 and XRCC1 genes and the risk of childhood acute lymphoblastic leukemia Arch Toxicol 86(3) 431–9 DOI: 10.1007/s00204-011-0760-8
50. Alves S et al (2002) The GSTM1 and GSTT1 genetic polymorphisms and susceptibility to acute lymphoblastic leukemia in children from north Portugal Leukemia 16(8) 1565–7 DOI: 10.1038/sj.leu.2402543 PMID: 12145701
51. Barnette P et al (2004) High-throughput detection of glutathione s-transferase polymorphic alleles in a pediatric cancer population Cancer Epidemiol Biomarkers Prev 13(2) 304–13 DOI: 10.1158/1055-9965.EPI-03-0178 PMID: 14973099
52. Chen CL et al (1997) Higher frequency of glutathione S-transferase deletions in black children with acute lymphoblastic leukemia Blood 89(5) 1701–7 PMID: 9057653
53. Klotz J et al (2006) Population-based retrieval of newborn dried blood spots for researching paediatric cancer susceptibility genes Paediatr Perinat Epidemiol 20(5) 449–52 DOI: 10.1111/j.1365-3016.2006.00749.x PMID: 16911024
54. Pigullo S et al (2007) Are genotypes of glutathione S-transferase superfamily a risk factor for childhood acute lymphoblastic leukemia? Results of an Italian case-control study Leukemia 21(5) 1122–4 PMID: 17315021
55. Wang J et al (2004) Genetic polymorphisms analysis of glutathione S-transferase M1 and T1 in children with acute lymphoblastic leukemia Journal of Huazhong Univ Sci Technolog Med Sci 24(3) 243–4 DOI: 10.1007/BF02832001
56. Gatedee J et al (2007) Glutathione S-transferase P1 genotypes, genetic susceptibility and outcome of therapy in thai childhood acute lymphoblastic leukemia Asian Pac J Cancer Prev 8(2) 294–6 PMID: 17696749
57. Krajinovic M, Labuda D and Sinnett D (2002) Glutathione S-transferase P1 genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukaemia Pharmacogenetics 12(8) 655–8 DOI: 10.1097/00008571-200211000-00010 PMID: 12439226
58. Krajinovic M et al (2000) Genetic polymorphisms of N-acetyltransferases 1 and 2 and gene-gene interaction in the susceptibility to childhood acute lymphoblastic leukemia Cancer Epidemiol Biomarkers Prev 9(6) 557–62 PMID: 10868688
59. Zanrosso CW et al (2012) Genetic variability in N-acetyltransferase 2 gene determines susceptibility to childhood lymphoid or myeloid leukemia in Brazil Leuk Lymphoma 53(2) 323–7 DOI: 10.3109/10428194.2011.619605
60. Zanrosso CW et al (2010) N-acetyltransferase 2 polymorphisms and susceptibility to infant leukemia with maternal exposure to dipyrone during pregnancy Cancer Epidemiol Biomarkers Prev 19(12) 3037–43 DOI: 10.1158/1055-9965.EPI-10-0508 PMID: 20884738
61. Hattori H et al (2007) Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia Leukemia Res 31(12) 1633–40 DOI: 10.1016/j.leukres.2007.04.009
62. Jamroziak K et al (2004) Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia Eur J Haematol 72(5) 314–21 DOI: 10.1111/j.1600-0609.2004.00228.x PMID: 15059065
63. Semsei AF et al (2008) Association of some rare haplotypes and genotype combinations in the MDR1 gene with childhood acute lymphoblastic leukaemia Leuk Res 32(8) 1214–20 DOI: 10.1016/j.leukres.2007.12.009 PMID: 18243305
64. Urayama KY et al (2007) MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia Cancer Epidemiol Biomarkers Prev 16(6) 1172–7 DOI: 10.1158/1055-9965.EPI-07-0007 PMID: 17548681
65. Pongstaporn W et al (2009) Polymorphism of glutathione S-transferase Omega gene: association with risk of childhood acute lymphoblastic leukemia J Cancer Res Clin Oncol 135(5) 673–8 DOI: 10.1007/s00432-008-0501-4
66. Liu CY et al (2008) Maternal and offspring genetic variants of AKR1C3 and the risk of childhood leukemia Carcinogenesis 29(5) 984–90 DOI: 10.1093/carcin/bgn071 PMID: 18339682 PMCID: 2902386
67. Batar B et al (2009) DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia Leuk Res 33(6) 759–63 DOI: 10.1016/j.leukres.2008.11.005
68. Canalle R et al (2011) Impact of thymidylate synthase promoter and DNA repair gene polymorphisms on susceptibility to childhood acute lymphoblastic leukemia Leuk Lymphoma 52(6) 1118–26 DOI: 10.3109/10428194.2011.559672 PMID: 21463130
69. Celkan T et al (2008) The difference between pre-B cell acute lymphoblastic leukemia and Burkitt lymphoma in relation to DNA damage repair gene polymorphisms in childhood Leuk Lymphoma 49(8) 1638–40 DOI: 10.1080/10428190802140063 PMID: 18608862
70. Chokkalingam AP et al (2011) Haplotypes of DNA repair and cell cycle control genes, X-ray exposure, and risk of childhood acute lymphoblastic leukemia Cancer Causes Control 22(12) 1721–30 DOI: 10.1007/s10552-011-9848-y PMID: 21987080 PMCID: 3206192
71. Joseph T et al (2005) DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia Cancer Lett 217(1) 17–24 DOI: 10.1016/j.canlet.2004.06.055
72. Meza-Espinoza JP et al (2009) XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia Genet Mol Res 8(4) 1451–8 DOI: 10.4238/vol8-4gmr687 PMID: 20013659
73. Pakakasama S et al (2007) Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia Pediatr Blood Cancer 48(1) 16–20 DOI: 10.1002/pbc.20742
74. Stanczyk M et al (2011) The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia Mol Biol Rep 38(1) 445–51 DOI: 10.1007/s11033-010-0127-x
75. Tumer TB et al (2010) DNA repair XRCC1 Arg399Gln polymorphism alone, and in combination with CYP2E1 polymorphisms significantly contribute to the risk of development of childhood acute lymphoblastic leukemia Leuk Res 34(10) 1275–81 DOI: 10.1016/j.leukres.2010.02.035 PMID: 20394984
76. Chrzanowska KH et al (2006) Carrier frequency of mutation 657del5 in the NBS1 gene in a population of Polish pediatric patients with sporadic lymphoid malignancies Int J Cancer 118(5) 1269–74 DOI: 10.1002/ijc.21439
77. Pastorczak A et al (2011) Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the Polish population Leuk Res 35(11) 1534–6 DOI: 10.1016/j.leukres.2011.07.034 PMID: 21889209
78. Mathonnet G et al (2003) Role of DNA mismatch repair genetic polymorphisms in the risk of childhood acute lymphoblastic leukaemia Br J Haematol 123(1) 45–8 DOI: 10.1046/j.1365-2141.2003.04551.x PMID: 14510941
79. Li Q et al (2011) hOGG1 Ser326Cys polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population Cancer Sci 102(6) 1123–7 DOI: 10.1111/j.1349-7006.2011.01928.x PMID: 21401806
80. Wang SL et al (2006) Polymorphisms in ERCC1 and susceptibility to childhood acute lymphoblastic leukemia in a Chinese population Leuk Res 30(11) 1341–5 DOI: 10.1016/j.leukres.2006.03.027 PMID: 16723154
81. Wu KH et al (2010) Significant association of XRCC4 single nucleotide polymorphisms with childhood leukemia in Taiwan Anticancer Res 30(2) 529–33 PMID: 20332465
82. Hou X et al (2005) Cyclin D1 gene polymorphism and susceptibility to childhood acute lymphoblastic leukemia in a Chinese population Int J Hematol 82(3) 206–9 DOI: 10.1532/IJH97.A10418 PMID: 16207592
83. Do TN et al (2009) TP53 R72P and MDM2 SNP309 polymorphisms in modification of childhood acute lymphoblastic leukemia susceptibility Cancer Genet Cytogenet 195(1) 31–6 DOI: 10.1016/j.cancergencyto.2009.05.009 PMID: 19837266
84. Liberzon E et al (2004) Germ-line ATM gene alterations are associated with susceptibility to sporadic T-cell acute lymphoblastic leukemia in children Genes Chromosomes Cancer 39(2) 161–6 DOI: 10.1002/gcc.10306
85. Sheng X et al (2013) TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children Carcinogenesis 34(1) 228–35 DOI: 10.1093/carcin/bgs325
86. Phillips CL et al (2010) MDM2 polymorphism increases susceptibility to childhood acute myeloid leukemia: a report from the Children’s Oncology Group Pediatr Blood Cancer 55(2) 248–53 DOI: 10.1002/pbc.22519 PMID: 20582981 PMCID: 2915901
87. Piwkham D et al (2011) Multilocus association of genetic variants in MLL, CREBBP, EP300, and TOP2A with childhood acute lymphoblastic leukemia in Hispanics from Texas Cancer Epidemiol Biomarkers Prev 20(6) 1204–12 DOI: 10.1158/1055-9965.EPI-11-0059 PMID: 21493871
88. Papaemmanuil E et al (2009) Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia Nat Genet 41(9) 1006–10 DOI: 10.1038/ng.430 PMID: 19684604
89. Trevino LR et al (2009) Germline genomic variants associated with childhood acute lymphoblastic leukemia Nat Genet 41(9) 1001–5 DOI: 10.1038/ng.432 PMID: 19684603 PMCID: 2762391
90. Sherborne AL et al (2010) Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk Nat Genet 42(6) 492–4 DOI: 10.1038/ng.585 PMID: 20453839 PMCID: 3434228
91. Yang W et al (2010) ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence Leukemia 24(4) 894–6 DOI: 10.1038/leu.2009.277 PMID: 20054350
92. Enciso-Mora V et al (2012) Common genetic variation contributes significantly to the risk of childhood B-cell precursor acute lymphoblastic leukemia Leukemia 26(10) 2212–5 DOI: 10.1038/leu.2012.89 PMID: 22456626
93. Orsi L et al (2012) Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE) Leukemia 26(12) 2561–4 DOI: 10.1038/leu.2012.148 PMID: 22660188
94. Healy J et al (2010) Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia Haematologica 95(9) 1608–11 DOI: 10.3324/haematol.2010.022459 PMID: 20460642 PMCID: 2930966
95. Prasad RB et al (2010) Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood Blood 115(9) 1765–7 DOI: 10.1182/blood-2009-09-241513 PMID: 20042726
96. Ross JA et al (2013) Genetic variants modify susceptibility to leukemia in infants: a Children’s Oncology Group report Pediatr Blood Cancer 60(1) 31–4 DOI: 10.1002/pbc.24131
97. Vijayakrishnan J et al (2010) Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence Leuk Lymphoma 51(10) 1870–4 DOI: 10.3109/10428194.2010.511356 PMID: 20919861
98. Xu H et al (2012) ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia J Clin Oncol 30(7) 751–7 DOI: 10.1200/JCO.2011.38.0345 PMID: 22291082 PMCID: 3295551
99. Han S et al (2010) Genome-wide association study of childhood acute lymphoblastic leukemia in Korea Leuk Res 34(10) 1271–4 DOI: 10.1016/j.leukres.2010.02.001 PMID: 20189245
100. Ellinghaus E et al (2012) Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia Leukemia 26(5) 902–9 DOI: 10.1038/leu.2011.302 PMCID: 3356560
101. Hosking FJ et al (2010) Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk Blood 115 4472–7 DOI: 10.1182/blood-2009-09-244483 PMID: 20231427
102. Infante-Rivard C et al (2000) Parental smoking, CYP1A1 genetic polymorphisms and childhood leukemia (Quebec, Canada) Cancer Causes Control 11(6) 547–53 DOI: 10.1023/A:1008976116512 PMID: 10880037
103. Infante-Rivard C et al (1999) Risk of childhood leukemia associated with exposure to pesticides and with gene polymorphisms Epidemiology 10(5) 481–7 DOI: 10.1097/00001648-199909000-00004 PMID: 10468419
104. Infante-Rivard C, Amre D and Sinnett D (2002) GSTT1 and CYP2E1 polymorphisms and trihalomethanes in drinking water: effect on childhood leukemia Environmental health perspectives 110(6) 591–3 DOI: 10.1289/ehp.02110591 PMID: 12055050 PMCID: 1240875
105. Infante-Rivard C et al (2002) Childhood acute lymphoblastic leukemia associated with parental alcohol consumption and polymorphisms of carcinogen-metabolizing genes Epidemiology 13(3) 277–81 DOI: 10.1097/00001648-200205000-00007 PMID: 11964928
106. Infante-Rivard C (2003) Diagnostic x rays, DNA repair genes and childhood acute lymphoblastic leukemia Health Phys 85(1) 60–4 DOI: 10.1097/00004032-200307000-00012 PMID: 12852472
107. Infante-Rivard C, Mathonnet G and Sinnett D (2000) Risk of childhood leukemia associated with diagnostic irradiation and polymorphisms in DNA repair genes Environ Health Perspect 108(6) 495–8 DOI: 10.1289/ehp.00108495 PMID: 10856021 PMCID: 1638130
108. Perera FP et al (1999) Molecular epidemiologic research on the effects of environmental pollutants on the fetus Environ Health Perspect 107(Suppl 3) 451–60 DOI: 10.1289/ehp.99107s3451 PMID: 10346993 PMCID: 1566212