Association between tumour necrosis factor-a polymorphism and cervical cancer in Lagos State, Nigeria
Sarah O John-Olabode1, Ifeoma C Udenze2, Adebola A Adejimi3, Obiefuna Ajie2 and Kehinde S Okunade4
1Department of Haematology and Blood Transfusion, College of Medicine, University of Lagos, PMB 12003, Lagos, Nigeria
2Department of Clinical Pathology, College of Medicine, University of Lagos, PMB 12003, Lagos, Nigeria
3Department of Community Health, College of Medicine, University of Lagos, PMB 12003, Lagos, Nigeria
4Department of Obstetrics and Gynaecology, College of Medicine, University of Lagos, PMB 12003, Lagos, Nigeria
Abstract
Background: The data on tumour necrosis factor-α (TNF-α) promoter gene polymorphism in the African population are relatively limited, especially in Nigerian women.
Objectives: This study aimed to determine the prevalence and allele distribution of three TNF-α promoter gene SNPs loci – rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) in women with cervical cancer (CC) and then evaluated the association between TNF-α SNPs and CC among women in Lagos, Nigeria.
Methods: This is a cross-sectional study of 75 unmatched human immunodeficiency virus (HIV)-infected and uninfected women with and without CC enrolled from October 2021 to January 2023 at the gynaecological oncology, cytology, adult HIV and blood donor clinics of the Lagos University Teaching Hospital. About 5 mL of peripheral blood was collected from each participant for total Deoxyribonucleic acid extraction, primer synthesis and genotyping. The probability of developing CC based on the given SNP genotype was expressed as an odds ratio (OR) with a 95% confidence interval. Allelic frequency deviations from Hardy–Weinberg equilibrium were calculated using chi-square, and the statistical significance level was considered as two-tailed and set at p ≤ 0.05.
Results: Our study found that TNF-α −1031 T>C polymorphism was significantly associated with increased CC risk in HIV-negative women (HIV+/CC-; OR = 1.4, 95%CI 0.23–8.42, p = 0.03 and HIV-/CC-; OR = 1.37, 95%CI 0.01–1.68, p = 0.03) while the −308A>G A allele was also significantly associated with CC in HIV-positive women (OR = 1.33, 95%CI = 0.23–7.75).
Conclusion: We observed that HIV-negative and HIV-positive women who carry the C allele of −1031T>C and the A allele of −308G>A TNF-a promoter gene loci, respectively, are more susceptible to CC. We were also able to show protective linkages for the minor allele of the three SNPs of interest suggesting the potential of TNF-a as a surrogate marker for CC screening in addition to human papillomavirus primary testing. Further studies are required to determine the association between host factors and TNF-a polymorphism to harness the diagnostic and therapeutic advantage these associations will provide in the management of CC.
Keywords: cervical cancer, Nigeria, screening, polymorphism, TNF-α
Correspondence to: Kehinde S Okunade
Email: sokunade@unilag.edu.ng
Published: 12/02/2025
Received: 26/07/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Cervical cancer (CC) is the most common gynaecological cancer with over half a million new cases and over a quarter of a million deaths reported globally in 2020 [1]. Despite significant advances in the diagnosis, treatment and prevention of CC, the attributable burden of disease due to CC remains high especially in low- and middle-income countries [2]. Indeed, recent data shows the highest number of new cases and mortality due to CC is in the sub-Saharan Africa region [1, 3].
The pathogenesis of CC has been closely linked with various risk factors, including environmental, lifestyle and host factors. Women living with human immunodeficiency virus (HIV) infection are more susceptible to CC [4, 5]. According to the World Health Organisation, HIV-positive women have a six-fold higher risk of developing CC compared to women without HIV, with approximately 5% of all CC cases attributed to HIV [3]. CC is regarded as one of the acquired immunodeficiency syndromes-defining cancers [6]. HIV causes immunosuppression which increases the risk of human papillomavirus (HPV) infection [7]. Women living with HIV are more likely to have HPV co-infection and this creates a vicious cycle with HIV-promoting HPV-induced cervical carcinogenesis [3]. The interaction between HIV, HPV and host factors has been recognised to be involved in the process of cervical carcinogenesis [7]. In association with the synergy between HIV and HPV, inherited genetic predisposition in the host also appears to play a significant role in the development of invasive CC with accumulating evidence suggesting that polymorphisms in key regulatory genes (including tumour suppressor and other immune-related genes) play an important role in the susceptibility to HPV infection [8–15].
Tumour necrosis factor-α (TNF-α) gene single nucleotide polymorphisms (SNPs) have been associated with increased susceptibility to CC and these polymorphisms appear to be concentrated in the gene promoter region [16, 17]. Located on human chromosome 6 (6p21.3) in the Human Leukocyte Antigen (HLA) region between the Class I HLA-B and the Class II HLA-DR loci, the TNF-α gene codes for the TNF-α protein [16, 17]. TNF-α is a pro-inflammatory cytokine implicated in the clearance of HPV in vivo through the downregulation of viral gene transcription and promoting the host’s inflammatory response to eradicate the HPV. Additionally, TNF-α promotes apoptosis in CC cells [18–20]. While TNF-α is recognised to be of some benefit in CC through its proapoptotic role, some evidence has linked SNPs in the TNF-α promoter genes with CC and other solid tumours such as breast cancer in various racial populations [21, 22]. While some studies have shown some association between TNF-α promoter gene polymorphism and CC, this remains controversial as other studies observed no such relationship [20–27]. SNPs rs361525 (−238 G>A), rs1799964 (−1031 T>C) and rs1800629 (−308 G>A) have been studied extensively and notably rs1800629 (−308) polymorphism has been associated with the development of other solid tumours such as breast cancer [24–28].
Taken together, studying the genetic variants in CC is of diagnostic and prognostic value. Identifying genetic variants will help in understanding the biology of CC and serve as potential biomarkers and therapeutic targets. Although, CC screening using cytology has improved early disease detection, including genetic screening will provide information for disease characterisation, especially in high-risk women and improve disease monitoring and cost-effectiveness of CC management. The data on TNF-α promoter gene polymorphism in the African population are relatively limited, especially in Nigerian women. This study aimed to determine the prevalence and allele distribution of three TNF-α promoter gene SNPs loci – rs361525 (−238 G>A), rs1799964 (−1031 T>C) and rs1800629 (−308 G>A) in women with CC and then evaluated the association between TNFα SNPs and CC among women in Lagos, Nigeria.
Materials and methods
Study design and setting
This is a cross-sectional study conducted between October 2021 and January 2023 at the gynaecological oncology, cytology, adult HIV and blood donor clinics of the Lagos University Teaching Hospital (LUTH). LUTH is the leading tertiary healthcare facility in Lagos, Southwest Nigeria. It primarily serves as a specialised referral center for both public and private hospitals in Lagos and the neighbouring Ogun and Oyo States.
Study population
Eligible participants were 100 unmatched HIV-infected and HIV-negative women with and without CC enrolled at the study clinics. Inclusion criteria were HIV-positive women with pathologically confirmed cervical squamous cell carcinoma enrolled from the gynaecological oncology and cytology clinics (case group) and otherwise healthy HIV-positive and HIV-negative women randomly selected from the HIV and donor clinics during the same study period (comparison group). Exclusion criteria included history of other malignancies, refusal of consent at enrolment or withdrawal of consent during the study,
Study procedure
After ascertaining eligibility and collecting relevant sociodemographic and clinical information from each participant, about 4–5 mLs of peripheral blood samples were collected into ethylene diamine tetra acetic acid bottles at enrolment for Deoxyribonucleic acid (DNA) analysis. Total DNA was extracted from the individual’s buffy coats using the Quick-DNA ™ MiniPrep kit from Zymo Research (ZymoResearchCorp. Irvine, CA, USA, Catalog number: D3024) according to the manufacturer’s instructions and then stored at −20°c. Total DNA quality was assessed using NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA) using a 1.7–2.0 optical density range for the genotyping. The final sample concentration was diluted to 50 ng/μL and sent to Inqaba Biotechnical Industries (Pty) Ltd. (Pretoria, South Africa) for primer synthesis and genotyping using the Agena MassArray system (Agena Bioscience San Diego, CA, USA) (Figure 1, Table 1).
Statistical analysis
Statistical analysis was performed using SPSS version 29.0 (IBM Corp., Armonk, NY, USA). The Shapiro-Wilk method was used to test quantitative variable normality. Continuous variables were expressed as mean ± standard deviation (SD), and categorical variables were expressed in percentages and frequencies. Significant differences between the group continuous variables were tested with the Student’s T-test and analysis of variance while Pearson’s chi-square and Fisher’s exact test were used to evaluate differences in genotype distribution between subgroups. The probability of developing CC based on the given SNP genotype was expressed as an odds ratio (OR) with a 95% confidence interval (95%CI) [29]. Allelic frequency deviations from Hardy–Weinberg equilibrium (HWE) were calculated using chi-square [30]. The statistical significance level was considered as two-tailed and set at p ≤ 0.05.
Results
Out of the 100 participants recruited for this study, only 75 participants' DNA samples met the quality criteria for genotyping. The participants were further stratified into subgroups, namely HIV negative with CC (HIV–/CC+; n = 13, 17.1%), HIV positive with CC (HIV+/CC+; n = 34, 44.7%), HIV positive without CC (HIV+/CC–; n = 17, 22.4%) and HIV negative without CC (HIV–/CC–; n = 11, 14.5%) (Figure 2).
Overall, the ages of participants with CC were significantly higher than that of their comparison groups (p < 0.01) but no significant differences were observed in patients ages at menarche and coitarche between the subgroups – HIV–/CC+ versus HIV+/CC+ (57.62 ± 13.35 versus 58 ± 9.67 years, p = 0.94) and HIV+/CC– versus HIV–/CC– (43.18 ± 9.31 versus 41.82 ± 7.74 years, p = 0.68). The key demographic data are summarised in Table 2.
Genotypes and allelic distribution of the studied TNFα SNPs
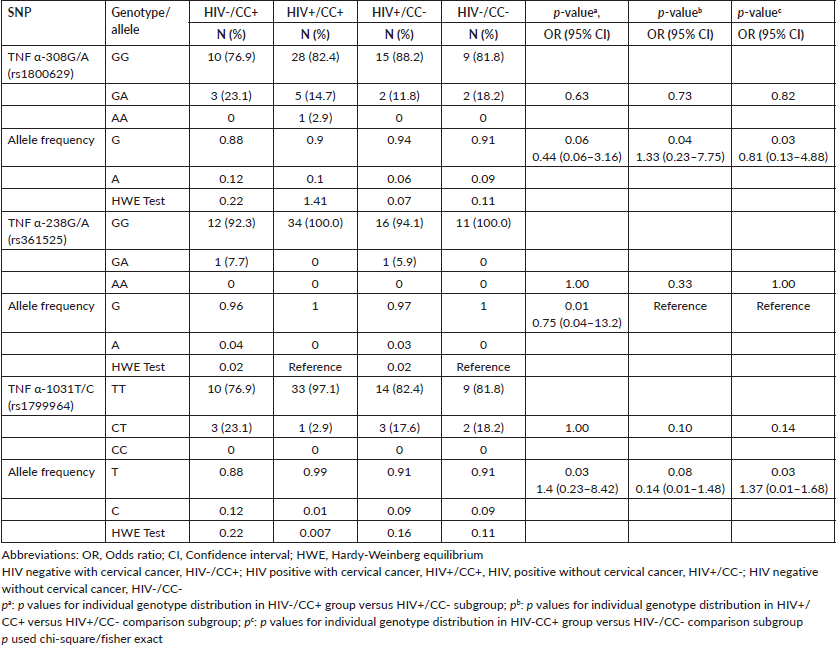
The allele distribution and genotype frequencies of −308/−238/−857 loci of TNF-α promoter among CC (with/without HIV) and the comparison groups are shown in Table 2. For TNF-α -308G>A, TNF-α −238G>A and TNF-α – 1031T>C SNPs, the genotype distributions were consistent with HWE in both the CC and comparison groups (Table 3).
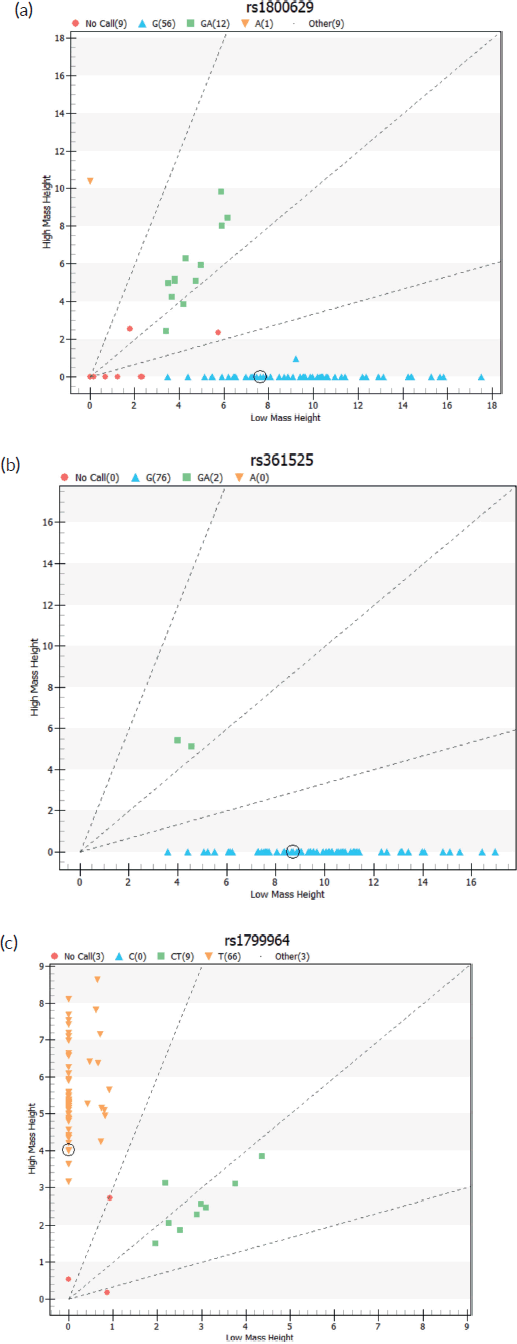
Figure 1. Cell cluster plots of TNF-α SNPs genotyping.
Table 1. Oligonucleotides probe and sequences.
TNF-α SNP genotyping test results by group
There was no significant difference in the TNF-α −308 homozygous and heterozygous genotype (GG/GA) between the CC cases and the comparators (pa = 0.63, pb = 0.73 and pc = 0.82, respectively, Table 3). We observed a higher distribution of the heterozygous GA genotype among the study group compared to the comparison group with 17% (8/47) in cases and 14.3% (4/28) in the comparators. Notably, the rare AA genotype was observed in the HIV+CC+ subgroup. The same was true in both the case and control groups for TNF-α -238(GG/GA) (pa = 1.00, pb = 0.33, respectively, and −1031(TT/CT) (pa =1.00, pb = 0.10 and pc = 0.14, respectively) (Table 3).
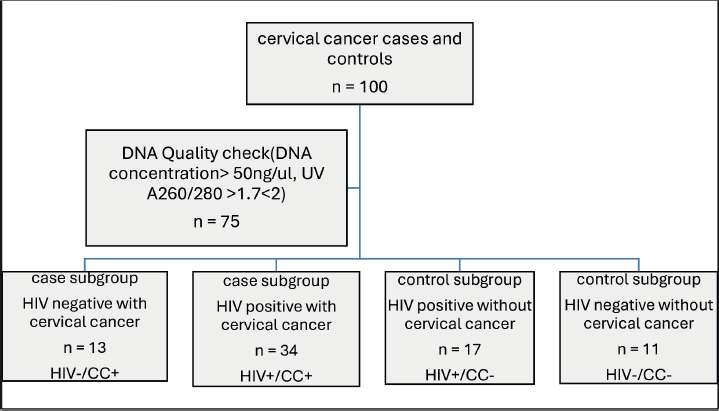
Figure 2: Flowchart of Participants’ stratification
Table 2. Demographics in the cases and comparison groups (n = 75).
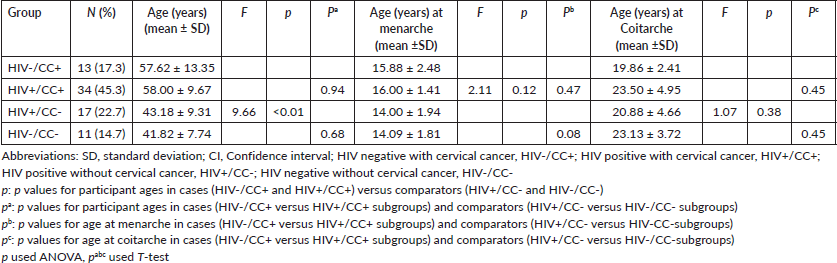
Table 3. Genotypes and allelic distribution of the studied TNFα SNPs.
Allele frequency of TNF-α −308G/A, −238G/A and −857C/T in CC and comparison groups
Regarding allele distribution of −308G>A, the A allele frequencies of −308G>A in the CC group HIV-/CC+ and HIV+/CC- in the comparison group were 12% and 6%, respectively, (OR = 0.44, 95%CI = 0.06–3.16, p = 0.06). The frequency of the A allele in the CC group HIV+/CC+ (10%) was significantly higher than the HIV+/CC- in the comparison group (6%) (p = 0.04). The estimated OR was 1.33 (95%CI = 0.23–7.75), which indicated an increased risk for the development of CC in HIV-positive women carrying the A allele of TNF α-308G>A. Comparing the HIV-/CC+ in the CC group and the HIV-/CC- in the comparison group (12% versus 9%), there was a significant difference in A allele frequency (OR = 0.81, 95%CI = 0.13–4.88, p = 0.03) but as observed previously there was no increased risk of CC in HIV negative women carrying the A allele of −308G>A (Table 3).
The A allele frequencies of −238G>A showed a significant difference between HIV-/CC+ in the CC group and HIV+/CC- in the comparison group (4% versus 3%), respectively (OR = 0.75, 95%CI = 0.04–13.2, p = 0.01). The OR<1 supports our observation of the A allele being a protective factor against CC in HIV-negative women. The HIV+/CC+ and HIV-/CC- comparison groups carried only the G allele (Table 3).
In Table 3, the C allele frequencies of −1031T>C of HIV-/CC+ in the CC group and comparison groups were significantly different (HIV+/CC-; OR = 1.4, 95%CI 0.23–8.42, p = 0.03 and HIV-/CC-; OR = 1.37, 95%CI 0.01–1.68, p = 0.03). The presence of the C allele had a significant association with CC in HIV-negative women (ORs = 1.4 and 1.37, respectively). In the HIV+CC+ and HIV+CC- subgroups, the C allele frequencies were 1% versus 9%, respectively, with OR<1 suggesting the presence of the C allele as a protective factor against CC in HIV-positive women (OR = 0.14, 95% CI 0.01–1.48 p = 0.08).
Discussion
Our study evaluated the association between genotype and allele frequency at three SNP loci of TNF-a promoter rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) and CC in Lagos State, Nigeria. We reported that the TNF-α −1031 T>C polymorphism was significantly associated with increased CC risk in HIV-negative women while the −308A>G A allele was also significantly associated with CC in HIV-positive women.
Contrary to previous studies [31, 32], our study revealed that the TNF-α −1031 T>C polymorphism was significantly associated with an increased risk of CC in HIV-negative women (OR = 1.4, 95%CI 0.23–8.42, p = 0.03). Our observations suggested that while HIV-negative women who carry the C allele in the TNF-α −1031 T>C gene locus are more susceptible to developing CC, the presence of the C allele offered protection from CC in HIV-positive women. Similar to evidence from the Asian population [21, 31, 33–35], in this study, the A allele of the −308G>A or −238G>A was found to be a protective factor for CC (OR = 0.72; 95% CI = 0.56–0.92 and OR = 0.75, 95%CI = 0.04–13.2) in HIV-negative women. Notably, the −308A>G A allele was found to be a risk factor for CC in HIV-positive women (OR = 1.33, 95%CI = 0.23–7.75). A review of previous literature reveals conflicting data on the association between −308G/A and −238G/A with CC in different or the same ethnic populations. While some studies conducted in Portuguese, Indian and Chinese women revealed an association between −308G>A and −238G>A and increased susceptibility to CC [16, 20, 36, 37], other studies carried out in women with diverse ethnicity including Mexican, Tunisian, American, South-African and Chinese populations reported the inheritance of the A allele of −308G>A or −238G>A and reduced the risk of developing CC [31, 33–35]. The possible explanations for the contradicting data in the different ethnic populations could be the diverse research methods used in these studies and most importantly the underlying host factors such as HIV infection. HIV infection causes a 6-fold rise in the risk of CC [3] due mostly to persistent HPV infection secondary to immune deficiency [38]. This has also been suggested by previous studies that reported that the host genetic background may facilitate HPV viral persistence in the uterine cervix as polymorphisms in coding regions of cytokine-like TNFA SNPs genes have been associated with susceptibility to some human diseases [14, 15]. However, we were not able to confirm this possible link in our present study as we did not assess the presence of genital high-risk HPV infection or its persistence in the enrolled participants.
TNF-α is one of the key inflammatory cytokines produced by macrophages that have been shown to play a crucial role in immune defense against pathogens including the control of HPV infection [21, 39]. TNF- α directly promotes the reduction of HPV gene transcripts, induces apoptosis in HPV infected/cancer cells and enhances inflammatory response to HPV. In addition to upregulating HPV presentation to effector T cells by antigen-presenting cells [40], evidence suggests TNF- α promoter region SNPs including rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) may cause deregulation of TNF- α transcription resulting in varied TNF- α levels in circulation. A low TNF- α results in reduced HPV antigen presentation with persistence of HPV infection while high TNF-α levels result in enhanced inflammatory response that drives cervical carcinogenesis with increased susceptibility to CC [32]. Our hypotheses in the study were – first, the genotype frequency of TNF-α promoter region SNPs is higher in HIV-negative women with CC than in comparators. Second, the genotype frequency of TNF-α promoter region SNPs is higher in HIV-positive women with CC than in HIV-negative women with CC and controls. The results we observed showed that the frequency of TNF-a promoter SNPs rs361525 (−238 G>A), rs1799964 (−1031T>C) and rs1800629 (−308 G>A) (7.7% versus 5.9%, 23.1% versus 18.2% and 23.1 versus 18.2%, p = 0.82, 1 and 0.14, respectively) in CC subgroup were not significantly different enough to reject the null hypothesis. In addition, our data were suggestive of the minor A allele of −308G>A and 238G>A giving a protective advantage for CC in HIV-negative women while −1031T>C minor C allele increased susceptibility for CC in the same group.
The frequency distribution of rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) alleles in the HIV-positive women with CC subgroup followed a similar pattern (0.0% versus 5.9%, 14.7% versus 11.8%, 2.9% versus 17.6%, p = 0.73, 0.33 and 0.10) with no statistically significant difference between the cases and comparators. In addition, our data were suggestive of the minor A allele of −1031T>C giving a protective advantage for CC in HIV-positive women while −308G>A minor A allele increased susceptibility for cervical in the same group. Our findings indicate the association of TNF-a promoter SNPs rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) with increased susceptibility to CC is dependent on underlying host factor, most especially factors that impair the immune system. This could be a possible reason for the conflicting findings in previous studies that have investigated the link between TNF-a promoter gene polymorphism and the risk of CC. It is, therefore, important to interpret data in the context of underlying host factors.
The main strength of this study is that is the first research, to the best of our knowledge, that evaluated the association between genotype and allele frequency at three SNP loci of TNF-a promoter rs361525 (−238 G>A), rs1799964(−1031 T>C) and rs1800629 (−308 G>A) and the risk of CC in Lagos State, Nigeria. The study, however, had several limitations. First, there was an age mismatch between the cases and comparison group thus making it probable that the protective advantage of SNPs of interest may have been overestimated as potential cases are currently classified as comparators. Second, the small sample size used in the study could potentially reduce the power to detect associations of minor alleles with CC. Third, the use of cervicovaginal washings instead of whole blood could have provided more information on the molecular dynamics within the cancer cells. Fourth, we did not investigate HPV infection; therefore, we could not directly examine any association between TNF-a promoter gene polymorphism and HPV viral persistence. Finally, there was no measurement of TNF-a serum level which could have given us an insight into the expression and functionality of the three TNF-a SNPs of interest. Based on these observations, we suggest that future research with a more robust sample size should be designed to validate the association between the identified TNF-a promoter gene polymorphisms and susceptibility to CC.
Conclusion
We observed that HIV-negative and HIV-positive women who carry the C allele of −1031T>C and the A allele of −308G>A TNF-a promoter gene loci, respectively, are more susceptible to CC. We were also able to show protective linkages for the minor allele of the three SNPs of interest and this would suggest the potential of TNF-a as a surrogate marker for CC screening, most especially in developing countries such as Nigeria where universal HPV vaccination, especially for school-age adolescents, is still lacking. This study is unique in its use of clearly defined case and comparison subgroups, focusing on a particularly vulnerable group of women. However, association studies are devoid of experimental evidence that establishes causation; therefore, further studies are required to determine the association between host factors and TNF-a polymorphism to harness the diagnostic and therapeutic advantage these associations will provide in managing CC.
Acknowledgments
The authors thank Dr Aron and other staff of Inqaba Biotechnical Industries (Pty) Ltd. (Pretoria, South Africa) for their help in preparing the DNA samples, and the analyses of TNF-a polymorphism. Finally, the authors appreciate all the participating women without whom this study would have been impossible.
Human ethics and consent to participate
This study was approved by the College of Medicine, University of Lagos Health Research Ethics Committee (Approval number: ADM/DCST/HREC/APP/4377). The study was conducted per the Declaration of Helsinki. Before enrollment, all study participants provided written informed consent, and strict confidentiality of participants' information was ensured during and after the completion of the study.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author (KSO) upon reasonable request.
Conflicts of interest
The authors declare no competing interests in the conduct and publication of the study in this article.
Funding
The study was supported through the funding provided by the University of Lagos Central Research Committee grant (CRC NO 2021/01). The author (KSO) protected time was supported through funding from the National Cancer Institute and Fogarty International Center of the National Institutes of Health under Award Numbers K43TW011930 and D43TW010934 and the Conquer Cancer International Innovation Grant under Project ID 2024IIG-2761200216. The content of this paper is solely the responsibility of the authors. It does not necessarily represent the official views of the University of Lagos Central Research Committee, National Cancer Institute, Fogarty International Center or the National Institutes of Health and the Conquer Cancer Foundation. The funders had no role in the conception, the decision to publish or the preparation of this manuscript.
Author contributions
SOJ-O, ICU and KSO contributed to the study's conception and design. Material preparation, data collection and analysis were performed by SOJ-O, AAA, OA and KSO. The first draft of the manuscript was written by SOJ-O and KSO, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
References
1. Sung H, Ferlay J, and Siegel RL, et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 71(3) 209–249 https://doi.org/10.3322/caac.21660 PMID: 33538338
2. Sherer MV, Kotha NV, and Williamson C, et al (2022) Advances in immunotherapy for cervical cancer: recent developments and future directions Int J Gynecol Cancer 32 281–287 https://doi.org/10.1136/ijgc-2021-002492 PMID: 35256414
3. Stelzle D, Tanaka LF, and Lee KK, et al (2021) Estimates of the global burden of cervical cancer associated with HIV Lancet Glob Health 9(2) e161–e169 https://doi.org/10.1016/S2214-109X(20)30459-9
4. Liu G, Sharma M, and Tan N, et al (2018) HIV-positive women have higher risk of human papilloma virus infection, precancerous lesions, and cervical cancer AIDS 32 795–808 https://doi.org/10.1097/QAD.0000000000001765 PMID: 29369827 PMCID: 5854529
5. Okunade KS, Badmos KB, and Soibi-Harry AP, et al (2023) Cervical epithelial abnormalities and associated factors among HIV-infected women in Lagos, Nigeria: a cytology-based study Acta Cytol 67(3) 248–256 https://doi.org/10.1159/000527905
6. Hernández-Ramírez RU, Shiels MS, and Dubrow R, et al (2017) Cancer risk in HIV-infected people in the USA from 1996 to 2012: a population-based, registry-linkage study Lancet HIV 4(11) e495–e504 https://doi.org/10.1016/S2352-3018(17)30125-X PMID: 28803888 PMCID: 5669995
7. Murenzi G, Tuyisenge P, and Kanyabwisha F, et al (2021) Type-specific persistence, clearance and incidence of high-risk HPV among screen-positive Rwandan women living with HIV Infect Agent Cancer 16 16 https://doi.org/10.1186/s13027-021-00355-6 PMID: 33608036 PMCID: 7893720
8. Ferenczy A and Franco E (2002) Persistent human papillomavirus infection and cervical neoplasia Lancet Oncol 3 11–16 https://doi.org/10.1016/S1470-2045(01)00617-9 PMID: 11905599
9. de Freitas AC, Gurgel AP, and Chagas BS, et al (2012) Susceptibility to cervical cancer: an overview Gynecol Oncol 126(2) 304–311 https://doi.org/10.1016/j.ygyno.2012.03.047 PMID: 22484226
10. Hildesheim A and Wang SS (2002) Host and viral genetics and risk of cervical cancer: a review Virus Res 89 229–240 https://doi.org/10.1016/S0168-1702(02)00191-0 PMID: 12445662
11. Chen D, Juko-Pecirep I, and Hammer J, et al (2013) Genome-wide association study of susceptibility loci for cervical cancer J Natl Cancer Inst 105 624–633 https://doi.org/10.1093/jnci/djt051 PMID: 23482656
12. Chen D and Gyllensten U (2015) Lessons and implications from association studies and post-GWAS analyses of cervical cancer Trends Genet 31(1) 41–54 https://doi.org/10.1016/j.tig.2014.10.005
13. Johanneson B, Chen D, and Enroth S, et al (2014) Systematic validation of hypothesis-driven candidate genes for cervical cancer in a genome-wide association study Carcinogenesis 35(9) 2084–2088 https://doi.org/10.1093/carcin/bgu125 PMID: 24879636
14. Wang SS, Gonzalez P, and Yu K, et al (2010) Common genetic variants and risk for HPV persistence and progression to cervical cancer PLoS One 5(1) e8667 https://doi.org/10.1371/journal.pone.0008667 PMID: 20084279 PMCID: 2801608
15. Wang SS, Bratti MC, and Rodriguez AC, et al (2009) Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer J Infect Dis 199 20–30 https://doi.org/10.1086/595563
16. Duarte I, Santos A, and Sousa H, et al (2005) G-308A TNF-alpha polymorphism is associated with an increased risk of invasive cervical cancer Biochem Biophys Res Commun 334(2) 588–592 https://doi.org/10.1016/j.bbrc.2005.06.137 PMID: 16009345
17. Shi Y, Li L, and Hu Z, et al (2013) A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12 Nat Genet 45 918–922 https://doi.org/10.1038/ng.2687 PMID: 23817570
18. Bequet-Romero M and López-Ocejo O (2000) Angiogenesis modulators expression in culture cell lines positives for HPV-16 oncoproteins Biochem Biophys Res Commun 277(1) 55–61 https://doi.org/10.1006/bbrc.2000.3628
19. Malejczyk J, Malejczyk M, and Köck A, et al (1992) Autocrine growth limitation of human papillomavirus type 16-harboring keratinocytes by constitutively released tumor necrosis factor-alpha J Immunol 149(8) 2702–2708 https://doi.org/10.4049/jimmunol.149.8.2702 PMID: 1328383
20. Tjiong MY, van der Vange N, and ter Schegget JS, et al (2001) Cytokines in cervicovaginal washing fluid from patients with cervical neoplasia Cytokine 14(6) 357–360 https://doi.org/10.1006/cyto.2001.0909 PMID: 11497498
21. Yang J, Wang Y, and Zhang S, et al (2022) The association of TNF-α promoter polymorphisms with genetic susceptibility to cervical cancer in a Chinese Han population Int J Gen Med 15 417–427 https://doi.org/10.2147/IJGM.S350263 PMCID: 8760922
22. Badano I, Stietz SM, and Schurr TG, et al (2012) Analysis of TNFα promoter SNPs and the risk of cervical cancer in urban populations of Posadas (Misiones, Argentina) J Clin Virol 53(1) 54–59 https://doi.org/10.1016/j.jcv.2011.09.030
23. Chinchai T, Homchan K, and Sopipong W, et al (2016) Lack of associations between TNF-αPolymorphisms and cervical cancer in Thai women Asian Pac J Cancer Prev 17(3) 953–956 https://doi.org/10.7314/APJCP.2016.17.3.953 PMID: 27039819
24. Duvlis S, Dabeski D, and Cvetkovski A, et al (2020) Association of TNF-a (rs361525 and rs1800629) with susceptibility to cervical intraepithelial lesion and cervical carcinoma in women from Republic of North Macedonia Int J Immunogenet 47(6) 522–528 https://doi.org/10.1111/iji.12506 PMID: 32662227
25. Shen C, Sun H, and Sun D, et al (2011) Polymorphisms of tumor necrosis factor-alpha and breast cancer risk: a meta-analysis Breast Cancer Res Treat 126(3) 763–770 https://doi.org/10.1007/s10549-010-1184-5
26. Tavares MC, de Lima Júnior SF, and Coelho AV, et al (2016) Tumor necrosis factor (TNF) alpha and interleukin (IL) 18 genes polymorphisms are correlated with susceptibility to HPV infection in patients with and without cervical intraepithelial lesion Ann Hum Biol 43(3) 261–268 https://doi.org/10.3109/03014460.2014.1001436
27. Jin Y (2015) Association of single nucleotide polymorphisms in tumor necrosis factor alpha with cervical cancer susceptibility Cell Biochem Biophys 71 77–84 https://doi.org/10.1007/s12013-014-0165-4
28. Li L, Liu J, and Liu C, et al (2018) The correlation between TNF-α-308 gene polymorphism and susceptibility to cervical cancer Oncol Lett 15(5) 7163–7167 PMID: 29725439 PMCID: 5920275
29. Rodriguez S, Gaunt TR, and Day IN (2009) Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies Am J Epidemiol 169(4) 505–514 https://doi.org/10.1093/aje/kwn359 PMID: 19126586 PMCID: 2640163
30. Bland JM and Altman DG (2000) Statistics notes. The odds ratio BMJ 320(7247) 1468 https://doi.org/10.1136/bmj.320.7247.1468 PMID: 10827061 PMCID: 1127651
31. Govan VA, Constant D, and Hoffman M, et al (2006) The allelic distribution of -308 tumor necrosis factor-alpha gene polymorphism in South African women with cervical cancer and control women BMC Cancer 6 24 https://doi.org/10.1186/1471-2407-6-24 PMID: 16438713 PMCID: 1397852
32. Li X , Yin G, and Li J, et al (2018) The correlation between TNF-α promoter gene polymorphism and genetic susceptibility to cervical cancer Technol Cancer Res Treat 17 1533033818782793 https://doi.org/10.1177/1533033818782793
33. Wang Y, Yang J, and Huang J, et al (2020) Tumor necrosis factor-alpha polymorphisms and cervical cancer: evidence from a meta-analysis Gynecol Obstet Invest 85(2) 153–158 https://doi.org/10.1159/000502955
34. Zidi S, Stayoussef M, and Zouidi F, et al (2015) Tumor necrosis factor alpha (-238 / -308) and TNFRII-VNTR (-322) polymorphisms as genetic biomarkers of susceptibility to develop cervical cancer among tunisians Pathol Oncol Res 21(2) 339–345 https://doi.org/10.1007/s12253-014-9826-2
35. Deshpande A, Nolan JP, and White PS, et al (2005) TNF-alpha promoter polymorphisms and susceptibility to human papillomavirus 16-associated cervical cancer J Infect Dis 191 969–976 https://doi.org/10.1086/427826 PMID: 15717274
36. Du GH, Wang JK, and Richards JR, et al (2019) Genetic polymorphisms in tumor necrosis factor alpha and interleukin-10 are associated with an increased risk of cervical cancer Int Immunopharmacol 66 154–161 https://doi.org/10.1016/j.intimp.2018.11.015 PMCID: 6348885
37. Singh H, Jain M, and Sachan R, et al (2009) Association of TNFA (−308G>A) and IL-10 (−819C>T) promoter polymorphisms with risk of cervical cancer Int J Gynecol Cancer 19(7) 1190–1194 https://doi.org/10.1111/IGC.0b013e3181a3a3af PMID: 19823053
38. Menon S, Wusiman A, and Boily MC, et al (2016) Epidemiology of HPV genotypes among HIV positive women in Kenya: a systematic review and meta-analysis PLoS One 11 e0163965 https://doi.org/10.1371/journal.pone.0163965 PMID: 27764092 PMCID: 5072621
39. Balkwill F (2006) TNF-alpha in promotion and progression of cancer Cancer Metastasis Rev 25(3) 409–416 https://doi.org/10.1007/s10555-006-9005-3 PMID: 16951987
40. Basile JR, Zacny V, and Munger K (2001) The cytokines tumor necrosis factor–α (TNF-α) and TNF-related apoptosis-inducing ligand differentially modulate proliferation and apoptotic pathways in human keratinocytes expressing the human papillomavirus–16 E7 oncoprotein J Biol Chem 276 22522–22528 https://doi.org/10.1074/jbc.M010505200 PMID: 11306566