Potential carcinogenic role of Reg IV in ulcerative colitis-associated colorectal neoplasia
Yosra Abdelmonem Zamzam1, Yomna Zamzam2,a, Ayman Elsaka2, Lamiaa Al Fadaly3, Tamer Haydara4 and Alaa Ibraheem Amer2
1Department of Clinical Pathology, Faculty of Medicine, Tanta University, Tanta 31111, Egypt
2Department of Pathology, Faculty of Medicine, Tanta University, Tanta 31111, Egypt
3Clinical Pathology, National Cancer Institute, Cairo University, Giza 12511, Egypt
4Department of Internal Medicine, Faculty of Medicine, Kafr El Sheikh University, Kafr El-Sheikh 33511, Egypt
ahttps://orcid.org/0000-0003-0270-3140
Abstract
Background: Early detection of ulcerative colitis-associated neoplasia (UC-N) remains a clinical challenge. Identification of molecular biomarkers for colorectal dysplasia and cancer may be extremely beneficial in early detection and managing cancer risk in long-standing ulcerative colitis (UC) patients.
Objective: The aim of this work is to investigate the role of Reg IV in comparison to P53 and KRAS in UC-associated dysplasia and colorectal cancer (CRC) in order to evaluate the potential use of Reg IV for dysplasia and cancer screening in UC patients.
Methods: The study was conducted on 5 groups each 20 colonic endoscopic samples: 1) Normal colonic mucosa, 2) Active UC without dysplasia/carcinoma, 3) UC-associated dysplasia, 4) UC-associated CRC (UC-CRC), 5) Sporadic CRC. All included cases were subjected to Reg IV mRNA expression analysis by quantitative reverse transcription polymerase chain reaction, and immunostaining for Reg IV, P53 and KRAS.
Results: Reg IV mRNA expression levels were found to be significantly higher in groups 3 and 4 (mean: 3.37 and 5.70, respectively). Reg IV immunostaining was highly expressed in groups 3 and 4 (mean: 45.80 and 62.35, respectively). While P53 and KRAS immunostaining was highly expressed in group 5 (mean: 64.57 and 62.90). Furthermore, Reg IV immunoexpression had shown a negative correlation with P53 and KRAS immunoexpression in groups 4 and 5.
Conclusion: Higher expression of Reg IV in patients with UC-dysplasia and UC-CRC versus KRAS and P53 expression in sporadic CRC, suggests a potential role of Reg IV in UC carcinogenesis pathway. This could advocate the use of Reg IV as a screening biomarker for UC-N among patients with long-standing UC as well as a promising targeted therapeutic strategy.
Keywords: ulcerative colitis, neoplasia, dysplasia, colorectal cancer, Reg IV, PCR
Correspondence to: Yomna Abdelmonem Zamzam
Email: yomnazamzam@hotmail.com
Published: 29/08/2024
Received: 10/04/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Ulcerative colitis (UC) is defined as a chronic relapsing inflammatory bowel disease that affects the colon and rectum, leading to continuous mucosal inflammation [1]. The incidence of UC-associated neoplasia (UC-N) including both dysplasia and cancer increases in patients who have long-standing and extensive UC. In order to improve the prognosis of UC-associated colorectal cancer (UC-CRC), it is significant to diagnose it at an early or precancerous state in patients with long-duration UC [2].
Although UC-CRC accounts for just 1%–2% of colorectal cancers (CRCs) in the general population, it contributes to nearly 15% of all deaths in UC patients. Thus, it is critical to identify high-risk patients and carry out the appropriate monitoring [3].
The pathogenesis of UC-CRC is considered a multifactorial process, starting from no dysplasia to irreversible dysplasia, followed by UC-associated dysplasia (UC-dysplasia) which is divided into low-grade and high-grade dysplasia leading eventually to carcinoma [4]. Various immunological and genetic alterations, including oncogene overexpression, tumor suppressor gene inactivation and mutations, are involved in the oncogenesis process [5].
A specific series of changes in tumor suppressor genes and oncogenes that are commonly observed in sporadic CRC (S-CRC) may be also significant in the carcinogenesis of UC-CRC [6]. TP53, a tumor suppressor gene and KRAS, a proto-oncogene, have been strongly implicated in S-CRC [7]. Nevertheless, involved gene sequences and mutation frequencies of TP53 and KRAS can be different between UC-CRC and S-CRC, their sequence of events may also differ. Instead of the typical adenoma-carcinoma sequence, UC-CRC develops with an inflammatory-dysplasia-carcinoma sequence [8].
Regenerating islet-derived family, member 4 (Reg IV) is a member of the Reg gene family, belonging to the calcium-dependent lectin superfamily [9]. Reg IV is a secretory protein that is expressed in various normal tissues, including the pancreas, small intestine, colon and stomach [10]. Increased expression of Reg IV has been found in inflammatory bowel diseases [11] and various cancerous tissues such as CRC [12], gastric cancer [13], prostate cancer [14] and pancreatic cancer [15].
For the early detection of dysplasia, it is crucial that the etiology of CRC development in UC patients can be understood. Moreover, understanding the differences in carcinogenesis between UC-CRC and S-CRC could offer a new tool for targeted therapy in patients with UC-CRC [16].
Colonoscopy is mainly recommended for surveillance; however, because of the inflammatory and regenerative changes in the colonic mucosa, endoscopic and histologic diagnosis of dysplasia is often challenging [17]. Consequently, an objective molecular biomarker for dysplasia and cancer screening would be beneficial for cancer risk management in UC patients.
In this study, we sought to investigate the role of Reg IV in comparison to P53 and KRAS in UC-associated dysplasia and CRC in order to evaluate the potential use of Reg IV for dysplasia and cancer screening in patients with long-standing UC.
Material and methods
Case selection
This prospective study was conducted from May 2021 to 2023, among Internal medicine, Pathology and Clinical Pathology Departments, Kafr El Sheikh University and Tanta University, Egypt.
During this period, endoscopic samples were selected from patients with long-standing UC more than 10 years either for follow-up or complaining of rectal bleeding, chronic diarrhea and abdominal pain. Besides, endoscopic samples from patients presented with clinically suspicious colonic masses without a history of UC or hereditary CRC were also included for S-CRC sample selection.
Sample collection
Two separate endoscopic samples were obtained from the same lesion in all included patients. One sample was for histopathological examination and the other for molecular study. After histopathological examination of colonoscopic tissue samples, the cases were divided into five groups each with 20 samples:
1) Normal colonic mucosal specimens (control) were obtained from the colonic mucosa which showed no evidence of histological inflammation.
2) Active UC without dysplasia/carcinoma group showed evidence of neutrophils in the lamina propria, crypt abscess and crypt distortion.
3) UC-dysplasia group. Dysplasia was defined as any of altered nuclear/cytoplasmic ratio, increased cell size and/or an increase in mitotic figures.
4) UC-CRC group
5) S-CRC group.
Groups 1 and 2 were considered non neoplastic cases and the other groups were neoplastic cases. The clinicopathological features such as age at time of operation, sex of the patients and site of the lesion, were collected from the medical records.
All included cases were subjected to quantitative reverse transcription polymerase chain reaction (qRT‐PCR) for Reg IV mRNA expression analysis and immunostaining for Reg IV, P53 and KRAS.
Inclusion criteria
1) Patients with long-standing UC more than 10 years for endoscopic surveillance, 2) UC patients complaining of rectal bleeding, chronic diarrhea or abdominal pain, 3) UC patients with clinically suspicious colonic mass, 4) individuals undergoing a colonoscopy to check for CRC and 5) patients ranging in age from 35 to 75 years old.
Exclusion criteria
Patients with any of the following were excluded from the study; 1) other type of colitis other than UC, 2) hereditary CRC including familial adenomatous polyposis or hereditary nonpolyposis CRC, 3) family history of CRC 4) use of nonsteroidal anti-inflammatory medicines in the last 3 months and 5) history of other malignant tumors, autoimmune disease or other chronic inflammatory disease.
Ethics approval and consent to participate
This study was approved by the ethics committee of the Faculty of Medicine, Tanta University (approval code, 36264PR492) in accordance with the Declaration of Helsinki for experiments in humans and written informed consent was obtained from each participant.
Histopathological evaluation
After colonoscopy, tissue samples obtained for histopathological examination were fixed in formalin, embedded as paraffin blocks and sectioned at 4 μm, and stained with eosin and hematoxylin.
Immunohistochemistry procedure
All slides were briefly rehydrated and antigen retrieval was carried out by sodium citrate (pH = 6.0) in a pressure cooker (EDTA buffer, pH = 8.4). Endogenous peroxidase combined with 3% hydrogen peroxide was used to block all slides, whereas 2.5% bovine serum albumin in phosphate-buffered saline was used to block non-specific protein.
The following primary antibodies were utilized: anti-REG IV (1:50; R&D Systems, Minneapolis, MN, USA), anti-P53 (1:50; clone DO-7, cat # M7001, Dako) and anti-KRAS mouse monoclonal (1:10; Clone-F234, Santacruz biotechnology) and counterstained with hematoxylin.
The degree of immunostaining of formalin-fixed, paraffin-embedded sections was scored and reviewed independently by three different pathologists. Cytoplasmic with or without nuclear localization for Reg IV, cytoplasmic localization KRAS and nuclear localization for P53 are considered positive. Staining was categorized as negative, score 0 (0-<10%) or positive and scored as the overall proportion of cells (score 1: 10%–50% and score 2: >50%]. Quantitative calculation in the area of maximum staining per 10 high power fields using image analysis software (image J software).
Quantitative reverse transcription polymerase chain reaction
Extraction of total RNA from frozen tissue samples by utilizing RNA extraction kits (RNeasy FFPE Kit, Qiagen, Germany) strictly following the instructions of manufacture. The concentrations of RNA were quantitatively measured at 260 nm absorbance using NanoDrop 2000/2000c Spectrophotometer (Thermo Scientific, USA). QuantiTect® Reverse Transcription (Qiagen, Germany) was conducted to reverse transcribe the isolated RNA to cDNA. Reg IV mRNA expression was relatively quantified using TaqMan assay kits (Thermo Scientific, USA).
The primers used in the study: for Reg IV: forward:5′-CAGATCCTGGTCTGGCAAGT-3′ and reverse: 5′- ATTCGTTGCTGCTCCAAGTT-3′. Internal control GAPDH was used for each sample (forward primer: 5′-ACCACAGTCCATGCCATCCAC-3′; reverse primer: 5′-TCCACCACCCTGTTGCTGTA-3′). The plate was applied on real-time PCR system (Applied Biosystems, Canada) including the following thermal profile: holding at 95°C for 10 seconds then 45 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 30 seconds. The cycle threshold (CT) was gained for the gene using Applied Biosystems, step I version, software analysis modules, Relative mRNA expression levels were determined using the 2−ΔΔCt method [18].
Statistical analysis
Data were fed to the computer and analyzed by IBM SPSS software package version 20.0. (Armonk, NY: IBM Corp). Categorical data were represented as percentages and numbers. A chi-square statistical test was used to compare categorical proportions. Quantitative data were expressed as mean, standard deviation, range (minimum and maximum) and median. One-way ANOVA test was used to compare the different studied groups. Kruskal Wallis test was used to compare different groups for not normally distributed quantitative variables. Correlation between two normally distributed quantitative variables was conducted by the Pearson coefficient. The receiver operating characteristic curve (ROC) was utilized to assess the sensitivity and specificity of quantitative diagnostic measures between different groups. The area under the curve (AUC) is the area between the curve and the reference line, it represents how the markers are capable of distinguishing between the studied groups.
Results
The clinico-pathological characteristics of the five studied groups were shown in Table 1. The predominant sex in all studied groups was males (50%–80%). Regarding location, the rectum and left colon were the most common sites for UC and UC-dysplasia groups (40% and 35%, respectively), the right colon for UC-CRC [45%] and the left colon for S-CRC (50%). The mean age of the studied groups varied between 52.0 years for group 2 and 55.30 years for group 4. There was a statistically significant difference among studied groups regarding location (p < 0.001); while there was no statistically significant difference regarding age and sex (p value: 0.464 and 0.353, respectively).
Regarding the molecular analysis of studied cases, Reg IV staining was highly expressed in groups 3 and 4 (mean 45.80 and 62.35, respectively). On the other hand, P53 and KRAS immunostaining were highly expressed in group 5 (mean, 64.57 and 62.90). Reg IV mRNA expression was significantly higher in groups 3 and 4 (mean 3.37 and 5.70, respectively). All these findings were statistically significant between the five studied groups (p value < 0.001). These findings were all summarized in Table 2 and Figure 1.
The number of stained cells was scored as previously described in to three scores (Figures 2–4). Higher staining scores for Reg IV expression were found in groups 3 and 4 while the higher staining scores for P53 and KRAS were found in group 5. Regarding group 1, the staining score of all studied markers was mainly of score 0 (90% for Reg IV and 100% for each P53 and KRAS). For group 2, the staining score of Reg IV was mainly of score 1 (55%) and score 0 for both P53 and KRAS (90% and 95%, respectively). For group 3, the staining score of Reg IV was mainly of score 2 (45%) and score 0 for both P53 and KRAS (85% and 90%, respectively). For group 4, the staining score of Reg IV was mainly of score 3 (85%) and score 0 for both P53 and KRAS (70% and 80%, respectively). For group 5, the staining score of Reg IV was mainly of score 0 (80%) and score 2 for both P53 and KRAS (90% and 85%, respectively).
All these findings were statistically significant between the five studied groups (p value < 0.001). These results are summarized in Table 3.
There was a negative correlation between Reg IV immunoexpression and P53 expression in both group 4 and group 5 (−0.537 and −0.106, respectively). Moreover, there was a negative correlation between Reg IV immunoexpression and KRAS in groups 4 and 5 (−0.792 and −0.189, respectively) as shown in Table 4.
As described before P53 and KRAS immunostaining was mainly negative (score 0) in groups 3 and 4 so, the ROC curve analysis was carried out to evaluate the validity of the Reg IV in discriminating between neoplastic tissue associated with UC (groups 3 + 4) from non-neoplastic colonic mucosa (groups 1 + 2) as illustrated in Table 5 and Figure 5. The sensitivity of Reg IV immunoexpression and mRNA expression was 87.5% and 92.5%, respectively. The specificity of Reg IV immunoexpression and mRNA expression was 85% and 90%, respectively. The cut off values for Reg IV and Reg IV mRNA expression were more than 17.93% and 2.01%, respectively.
Table 1. Clinico-pathological characteristics in the five studied groups.
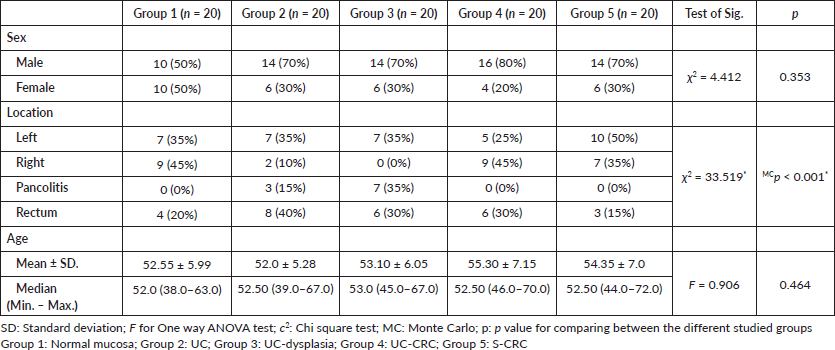
Table 2. Immunostaining of studied markers and Reg IV mRNA expression (molecular analysis) in between the five studied groups.
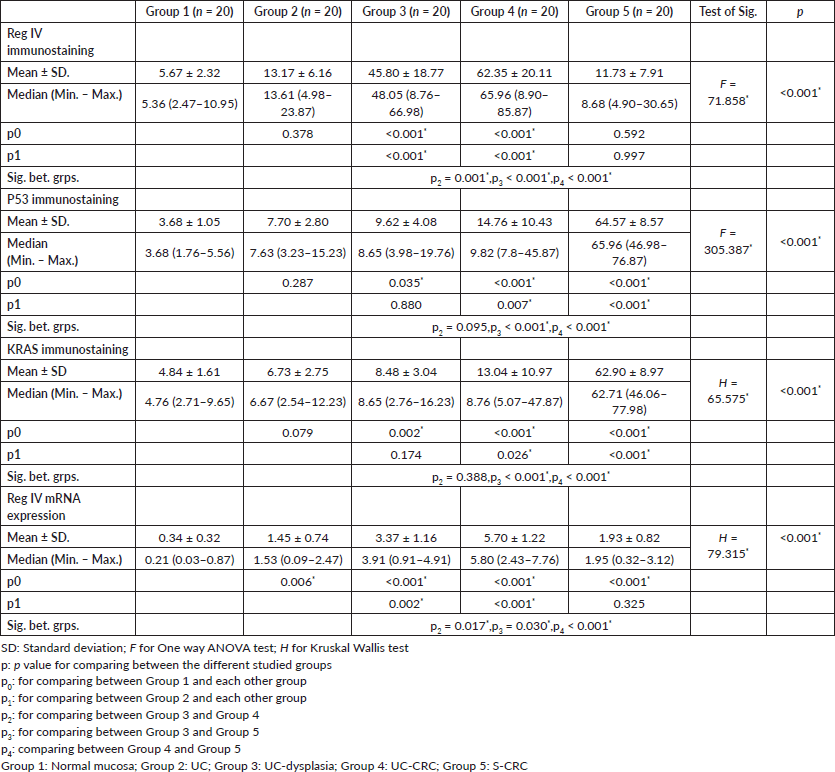
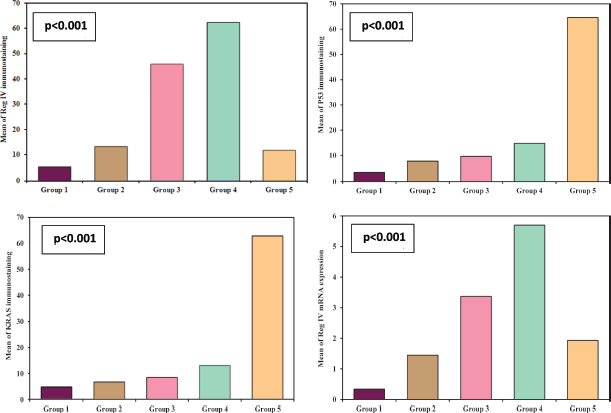
Figure 1. Immunostaining of studied markers and Reg IV mRNA expression in between the five studied groups.
Table 3. The score of Reg IV, P53 and KRAS immunostaining among the five studied groups.
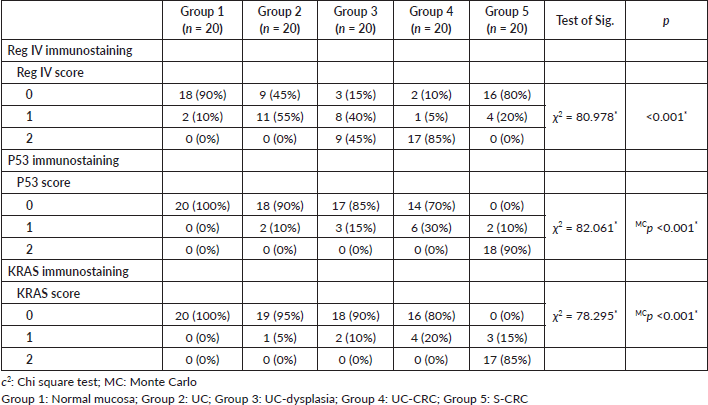
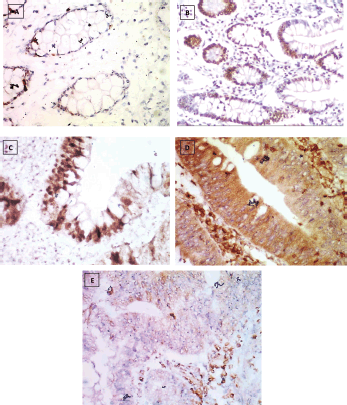
Figure 2. Reg IV immunostaining, (a): Negative expression in normal mucosa (group 1) 0.87% score 0, (b): Positive expression in UC case (group 2) 43.61% score 1, (c): Positive expression in UC-dysplasia case (group 3) 58.46% score 2, (d): Positive expression in UC-CRC case (group 4) 82.23% score and (e): Negative expression in S-CRC case (group 5) 6.15% score 0 (×400).
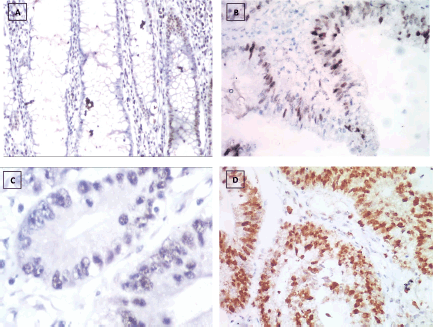
Figure 3. P53 immunostaining, (a): Negative expression in UC case (group 2) 2.45% score 0, (b): Positive expression in UC-dysplasia case (group 3) 26.18% score 1, (c): Negative expression in UC-CRC case (group 4) 3.47% score 0 and (d): Positive expression in S-CRC case (group 5) 76.34% score 2 (×400).
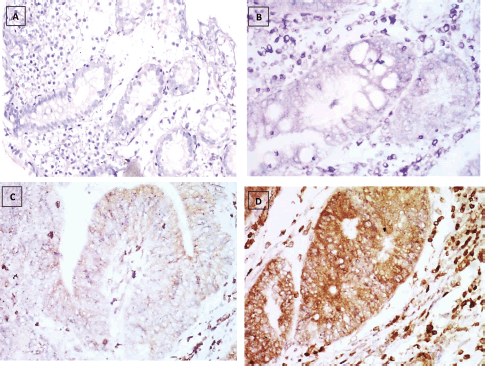
Figure 4. KRAS immunostaining, (a): Negative expression in UC case (group 2) 0.74% score 0, (b): Negative expression in UC-dysplasia case (group 3) 2.81% score 0, (c): Negative expression in UC-CRC case (group 4) 7.34% score 0 and (d): Positive expression in S-CRC case (group 5) 81.04% score 2 (×400).
Table 4. Correlation between Reg IV versus P53 and KRAS in each group.
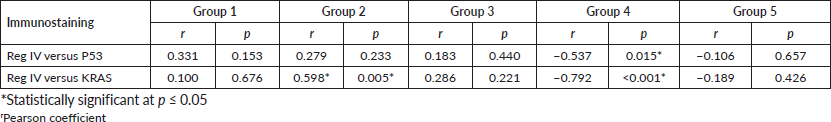
Table 5. Diagnostic performance for Reg IV to discriminate between group 3 + 4 (n = 40) from group 1 + 2 (n = 40).
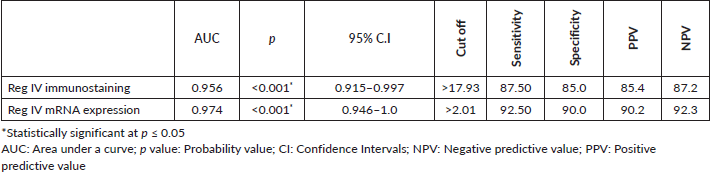
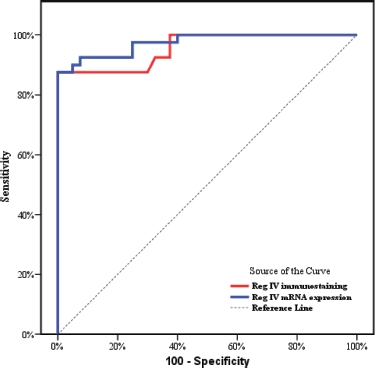
Figure 5. ROC curve for Reg IV immunostaining and mRNA expression to discriminate group 3 + 4 (n = 40) from group 1 + 2 (n = 40).
Discussion
UC-CRC is frequently diagnosed at an advanced stage and due to the higher mortality rate and worse prognosis in UC-CRC compared to S-CRC, early prediction of UC-CRC is crucial [4]. Although traditional endoscopic screening has been conducted for years, its efficiency primarily depends on operator experience and improvements in the endoscopic facility. Hence, molecular monitoring have an enormous potential to enhance the clinical management of the UC-N [3].
In the current study, our results demonstrated that Reg IV mRNA and immunohistochemical expressions in colorectal tissue were significantly elevated in UC-dysplasia and UC-CRC patients in comparison to normal colorectal tissue, UC and S-CRC groups, which may suggest the significance of Reg IV in the development of colitic cancer from UC mucosa. These findings were consistent with Nanakin et al [19] who revealed that REG IV mRNA was strongly enhanced in dysplasia and cancerous lesions in UC tissues. Previous studies have also reported the overexpression of Reg IV gene in colorectal adenocarcinoma, gastric cancer and pancreatic cancer [12, 13, 15].
Reg IV may be involved in colitic cancer development through its growth-promoting action. The basic biological functions of Reg family proteins include the induction of cell proliferation, cell migration, cell growth and cell adhesion [20]. Reg IV protein was reported to have antiapoptotic as well as mitogenic effects on colon cancer cells, via activating Akt signaling [19]. Therefore, Reg IV may play a mitogenic and/or antiapoptotic role in CRC development.
In vitro studies have revealed that Reg IV expression was induced by hepatocyte growth factor and basic fibroblast growth factor via the MAPK-dependent pathway. Moreover, colon cancer cells overexpressing REG IV gained significant growth ability [21]. Taken together, these findings imply that REG IV may promote epithelial cell proliferation in the UC-colitic cancer sequence.
Reg IV may also promote carcinogenesis by activating the epidermal growth factor receptor (EGFR), as Reg IV is a strong stimulator of the EGFR/Akt/activator protein-1 (AP-1) signaling pathway in colon cancer cells [22]. Therefore, suppressing endogenous Reg IV expression or inhibiting Reg IV/EGFR signaling may have the potential as a novel therapeutic tool to prevent dysplasia and cancer associated with UC.
In this study, P53 and KRAS immunostaining were highly expressed in S-CRC patients compared to UC-CRC. This was in agreement with past studies [23, 24]. Furthermore, Reg IV immunoexpression had shown a negative correlation with P53 and KRAS immunoexpression in UC-CRC and S-CRC groups. As demonstrated in many studies S-CRC develops with a sequence of molecular events, including alterations of KRAS, p53, adenomatous polyposis coli; however, molecular events leading to carcinogenesis in UC-CRC and S-CRC may be different [25, 26].
On the contrary, UC-CRC develops through an inflammation–dysplasia–carcinoma pathway. The longer the duration of UC is, the greater the risk of CRC [27]. This could be explained by the fact that prolonged inflammation can cause immunological dysfunction, the release of inflammatory cytokines and epigenetic alterations, all of which can result in dysplasia and cancer. This highlights the critical role that chronic inflammation plays in the pathogenesis of UC-CRC [28].
From the aforementioned results, it can be implied that targeting the KRAS/p53 pathway may not be an effective therapy in UC-CRC as opposed to S-CRC. However, Reg IV targeting could be a potential therapeutic tool in preventing dysplasia and cancer associated with UC.
ROC curve analysis was applied to assess the power Reg IV expression in differentiating UC-dysplasia and colitic cancer from normal colonic mucosa and UC groups. According to the ROC curve in our study, the sensitivity of Reg IV mRNA expression and immunostaining were 92.50% and 87.50%, respectively, and the specificity was 90.0% and 85.0%, respectively. The cut off of Reg IV mRNA relative expression and immunostaining in predicting dysplasia and colitic cancer in UC patients was found to be >2.01 and >17.93, respectively.
Our study revealed that Reg IV was up-regulated in UC-CRC tissues than in adjacent normal mucosa indicating that Reg IV overexpression may be an early event in colorectal carcinogenesis in long-standing UC patients. Most studies [11, 12, 19, 20] as well as the current study estimated Reg IV levels by immunohistochemistry staining and qRT‐PCR in tissue specimens, additionally Oue et al [29] used the enzyme-linked immunosorbent assay for Reg IV serum levels. Testing for Reg IV in the blood can potentially detect its level throughout the body; however, it does not indicate its definitive relation with UC or carcinoma in the colonic mucosa. Therefore, Reg IV expression should be investigated in colonic mucosa tissue samples for proper diagnosis [12, 19].
In conclusion, our results suggest that UC-dysplasia, UC-CRC and S-CRC may have different molecular pathways given the higher expression of Reg IV but lower KRAS and P53 expression in patients with UC-CRC in comparison to S-CRC. Our findings indicate the promising role of Reg IV as candidate biomarker for dysplasia and cancer screening in patients with UC. Hopefully, understanding the molecular pathway unique to UC-CRC may provide a useful tool for targeted therapy either by reducing endogenous Reg IV expression or blocking Reg IV downstream signaling. Ultimately, further clinical larger scale studies are needed to validate our findings.
Limitation of the study
There are some limitations in our study; the sample size is relatively small; therefore, future studies with bigger sample sizes are recommended to corroborate the current experimental results and to validate more anti-cancer agents that could target the Reg IV/EGFR/Akt/AP-1 signaling pathway.
Conflicts of interest
There is no conflict of interest.
Financial support and sponsorship
Nil.
References
1. Segal JP, LeBlanc JF, and Hart AL (2021) Ulcerative colitis: an update Clin Med (Lond) 21(2) 135–139 https://doi.org/10.7861/clinmed.2021-0080 PMID: 33762374 PMCID: 8002778
2. Buchman AL (2020) Ulcerative colitis: where we are and where we are not in 2020 Gastroenterol Clin North Am 49(4) xiii–xiv PMID: 33121698
3. Nishio M, Hirasawa K, and Chiba S, et al (2023) Endoscopic resection is feasible for high-grade dysplasia in patients with ulcerative colitis Scand J Gastroenterol 58(1) 101–106 https://doi.org/10.1080/00365521.2022.2107878
4. Kawachi H (2019) Histopathological diagnosis of ulcerative colitis-associated neoplasia Dig Endosc 31(1) 31–35 https://doi.org/10.1111/den.13387 PMID: 30994228
5. Karjalainen EK, Renkonen-Sinisalo L, and Lepistö AH (2020) Dysplasia in the mucosal biopsy specimen is still a warning sign of cancer in ulcerative colitis Scand J Gastroenterol 55(9) 1019–1023 https://doi.org/10.1080/00365521.2020.1794024 PMID: 32672485
6. Liu ZY, Zheng M, and Li YM, et al (2019) RIP3 promotes colitis-associated colorectal cancer by controlling tumor cell proliferation and CXCL1-induced immune suppression Theranostics 9(12) 3659–3673 https://doi.org/10.7150/thno.32126 PMID: 31281505 PMCID: 6587173
7. Patra R, Dey AK, and Mukherjee S (2023) Identification of genes critical for inducing ulcerative colitis and exploring their tumorigenic potential in human colorectal carcinoma PLoS One 18(8) e0289064 https://doi.org/10.1371/journal.pone.0289064 PMID: 37535606 PMCID: 10399749
8. Du L, Kim JJ, and Shen J, et al (2017) KRAS and TP53 mutations in inflammatory bowel disease-associated colorectal cancer: a meta-analysis Oncotarget 8(13) 22175–22186 https://doi.org/10.18632/oncotarget.14549 PMID: 28077799 PMCID: 5400656
9. Sun C, Wang X, and Hui Y, et al (2021) The potential role of REG family proteins in inflammatory and inflammation-associated diseases of the gastrointestinal tract Int J Mol Sci 22(13) 7196 https://doi.org/10.3390/ijms22137196 PMID: 34281249 PMCID: 8268738
10. Takasawa S, Itaya-Hironaka A, and Makino M, et al (2022) Upregulation of Reg IV and Hgf mRNAs by intermittent hypoxia via downregulation of microRNA-499 in cardiomyocytes Int J Mol Sci 23(20) 12414 https://doi.org/10.3390/ijms232012414 PMID: 36293268 PMCID: 9603944
11. Tsuchida C, Sakuramoto-Tsuchida S, and Taked M, et al (2017) Expression of REG family genes in human inflammatory bowel diseases and its regulation Biochem Biophys Rep 12 198–205 PMID: 29090282 PMCID: 5655384
12. Hu Y, Pan C, and Hu J, et al (2015) The role of Reg IV in colorectal cancer, as a potential therapeutic target Contemp Oncol (Pozn) 19(4) 261–264 PMID: 26557771 PMCID: 4631303
13. Zhang N, Chai D, and Du H, et al (2018) Expression of Reg IV and SOX9 and their correlation in human gastric cancer BMC Cancer 18(1) 344 https://doi.org/10.1186/s12885-018-4285-x PMID: 29587675 PMCID: 5870489
14. Gu Z, Rubin MA, and Yang Y, et al (2005) Reg IV: a promising marker of hormone refractory metastatic prostate cancer Clin Cancer Res 11(6) 2237–2243 https://doi.org/10.1158/1078-0432.CCR-04-0356 PMID: 15788672
15. He XJ, Jiang XT, and Ma YY, et al (2012) REG4 contributes to the invasiveness of pancreatic cancer by upregulating MMP-7 and MMP-9 Cancer Sci 103(12) 2082–2091 https://doi.org/10.1111/cas.12018 PMID: 22957785 PMCID: 7659226
16. Li W, Zhao T, and Wu D, et al (2022) Colorectal cancer in ulcerative colitis: mechanisms, surveillance and chemoprevention Curr Oncol 29(9) 6091–6114 https://doi.org/10.3390/curroncol29090479 PMID: 36135048 PMCID: 9498229
17. Yashiro M (2014) Ulcerative colitis-associated colorectal cancer World J Gastroenterol 20(44) 16389–16397 https://doi.org/10.3748/wjg.v20.i44.16389 PMID: 25469007 PMCID: 4248182
18. Livak KJ and Schmittgen TD (2021) Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCT method Methods 25 402–408 https://doi.org/10.1006/meth.2001.1262
19. Nanakin A, Fukui H, and Fujii S, et al (2007) Expression of the REG IV gene in ulcerative colitis Lab Invest 87(3) 304–314 https://doi.org/10.1038/labinvest.3700507 PMID: 17260007
20. Hartupee JC, Zhang H, and Bonaldo MF, et al (2001) Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: Reg IV Biochim Biophys Acta 1518(3) 287–293 https://doi.org/10.1016/S0167-4781(00)00284-0 PMID: 11311942
21. Bishnupuri KS, Luo Q, and Murmu N, et al (2006) Reg IV activates the epidermal growth factor receptor/Akt/AP-1 signaling pathway in colon adenocarcinomas Gastroenterology 130(1) 137–149 https://doi.org/10.1053/j.gastro.2005.10.001 PMID: 16401477
22. Cleveland NK and Rubin DT (2021) Cancer prevention in patients with inflammatory bowel disease Pract Gastroenterol 45(8) 12–28 PMID: 34707325 PMCID: 8547793
23. Carethers JM and Jung BH (2015) Genetics and genetic biomarkers in sporadic colorectal cancer Gastroenterology 149(5) 1177–1190.e3 https://doi.org/10.1053/j.gastro.2015.06.047 PMID: 26216840 PMCID: 4589489
24. Fabišíková K, Behulová RL, and Repiska V (2019) Molecular biomarkers in the diagnostic of patients with colorectal cancer Neuro Endocrinol Lett 40(5) 215–221
25. Hagland HR, Berg M, and Jolma IW, et al (2013) Molecular pathways and cellular metabolism in colorectal cancer Dig Surg 30(1) 12–25 https://doi.org/10.1159/000347166 PMID: 23595116
26. Nguyen HT and Duong HQ (2018) The molecular characteristics of colorectal cancer: implications for diagnosis and therapy Oncol Lett 16(1) 9–18 PMID: 29928381 PMCID: 6006272
27. Yin Y, Wan J, and Yu J, et al (2003) Molecular pathogenesis of colitis-associated colorectal cancer: immunity, genetics, and intestinal microecology Inflamm Bowel Dis 29(10) 1648–1657 https://doi.org/10.1093/ibd/izad081
28. Porter RJ, Arends MJ, and Churchhouse AMD, et al (2021) Inflammatory bowel disease-associated colorectal cancer: translational risks from mechanisms to medicines J Crohns Colitis 15(12) 2131–2141 https://doi.org/10.1093/ecco-jcc/jjab102 PMID: 34111282 PMCID: 8684457
29. Oue N, Kuniyasu H, and Noguchi T, et al (2007) Serum concentration of Reg IV in patients with colorectal cancer: overexpression and high serum levels of Reg IV are associated with liver metastasis Oncology 72(5-6) 371–380 https://doi.org/10.1159/000113147






