Epidermal growth factor receptor (EGFR) expression in the serum of patients with triple-negative breast carcinoma: prognostic value of this biomarker
Rogério Agenor de Araújo1,2a, Felipe Andrés Cordero da Luz2b, Eduarda da Costa Marinho2c, Camila Piqui Nascimento2d, Lara de Andrade Marques2e, Patrícia Ferreira Ribeiro Delfino2f, Rafael Mathias Antonioli2g, Breno Jeha Araújo3h, Ana Cristina Araújo Lemos da Silva1i, Maria Luiza Gonçalves dos Reis Monteiro1j, Morun Bernardino Neto4k and Marcelo José Barbosa Silva5l
1Federal University of Uberlândia, Avenida Pará, Bloco 2U, 1720, Campus Umuarama, Uberlândia, MG, CEP 38400-902, Brazil
2Cancer Research and Prevention Nucleus, Grupo Luta Pela Vida, Cancer Hospital in Uberlândia, Uberlândia, MG, CEP 38405-302, Brazil
3São Paulo State Cancer Institute of the Medical School of the University of São Paulo, São Paulo, SP, CEP 38405-302, Brazil
4Department of Basic and Environmental Sciences, University of São Paulo, Lorena, SP, CEP 12602-810, Brazil
5Laboratory of Tumor Biomarkers and Osteoimmunology, Institute of Biomedical Sciences, Federal University of Uberlândia, Uberlândia, MG, CEP 38405-320, Brazil
This study was published as an online abstract at the 2021 American Society of Clinical Oncology (ASCO) Annual Meeting.
ahttps://orcid.org/0000-0003-4653-6786
bhttps://orcid.org/0000-0002-9381-4913
chttps://orcid.org/0000-0002-1307-9104
dhttps://orcid.org/0000-0002-0955-8559
ehttps://orcid.org/0000-0002-2734-8352
fhttps://orcid.org/0000-0002-2196-9318
ghttps://orcid.org/0000-0003-3886-1562
hhttps://orcid.org/0000-0003-4892-9911
ihttps://orcid.org/0000-0002-8220-938X
jhttps://orcid.org/0000-0002-7961-6996
khttps://orcid.org/0000-0003-4292-7800
lhttps://orcid.org/0000-0002-5807-4286
Abstract
Background: Epidermal growth factor receptor (EGFR) overexpression has been considered a poor prognostic factor in breast cancer.
Methodology: A prospective study of 206 women with breast cancer analysed by stages (I, II, III and IV) and by immunohistochemical subtype (Luminal A, Luminal B, HER2+ and triple-negative (TN)); 89 healthy controls with normal recent mammography were included. The EGFR measured in the serum (sEGFR) was detected by the Enzyme-Linked Immunosorbent Assay (ELISA) method (R&D Systems kit DY231) collected by blood before any treatment in patients. Kaplan–Meier method and Cox regression were carried out to obtain the prognostic value, considering significance if p < 0.05.
Results: With a median follow-up of 36.6 months, 47 deaths occurred. Multivariable Cox regression showed difference of overall survival (OS) associated with sEGFR levels (sEGFR ≤ or > 47.8 ng/mL) in patients with TN cancers, but not of Luminal A, Luminal B or HER2+ subtypes; adjusted by stage, the death risk increased by approximately 415% [hazard ratio (HR): 5.149 (1.900–13.955), p = 0.001] for patients with sEGFR > 47.8 ng/mL compared to patients with a lower sEGFR value. There was no significant correlation of sEGFR with staging, histological tumour grade (G1/G2/G3), Ki67 (< or ≥14%) or body mass index.
Conclusions: Increased sEGFR expression in patients with TN tumours is a significant predictor of lower OS and its quantification is inexpensive and straightforward.
Keywords: breast cancer, prognosis, sEGFR, tEGFR
Correspondence to: Rogério A de Araújo
Email: rogeriodearaujo@ufu.br
Published: 20/07/2022
Received: 09/03/2022
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Breast cancer has a high mortality rate [1], but the prognosis can be improved if early diagnosis is made possible by mammography in older women [2], or by magnetic resonance imaging [3] in younger women [4]. After histological confirmation, TNM staging is performed and tumour subtypes are defined by immunohistochemical (IH), with hormone therapy for patients with luminal tumours [5], and monoclonal antibodies in those with Human Epidermal Receptor type 2 (HER2) overexpression [6], in addition to chemotherapy. However, for patients with triple-negative (TN) tumours, there are far fewer therapeutic options [5]. There are risks of recurrence at all subtypes, even in the early stages [7], requiring continuous treatment improvements [8].
Epidermal growth factor receptor (EGFR) is a glycoprotein, expressed as a transmembrane tyrosine kinase receptor, is studied as a biomarker in TN breast tumours, which can be expressed in 200,000–400,000 receptors in malignant cells. The EGFR has a half-life of 10 hours, undergoing intense recycling due to endocytosis, mainly by clathrin [9] and can be internalised in the cell nucleus. EGFR acts on gene transcription, increasing proliferation, reducing DNA damage by chemotherapy, and thus favouring the resistance to treatment [10]. EGFR can be measured by IH, Fluorescence In Situ Hybridization (FISH) or real-time C-reactive protein (RT-PCR) [11], although its intense turnover can hamper these detections.
TN tumours express other biomarkers such as p53, c-kit, claudin7, CK5/6, CK17, androgen receptors, PTEN, ALK and PDL-1 [12], demonstrating their heterogeneity [13]. The EGFR has a greater diversity of action, acting in the mesenchymal transformation of the epithelial cell [14], in addition to activating several signalling pathways with PI3K when dimerising with HER3, or with RAS when it binds to Grb2 proteins, or JAK when it binds to STAT proteins [15]. Among human epidermal receptors, only EGFR signals with RANK to up-regulate the AKT and ERK pathways [16]. EGFR is activated by EGF, which at a high concentration intensely internalises EGFR. In this condition, the cell adds ubiquitin molecules to EGFR [17]. This signalling can occur as a monomer or dimer with other ligands, such as amphiregulin present in extracellular vesicles, providing resistance to degradation [18]. The wild EGFR can signal through a non-canonical pathway [19], or even with chemokines such as CXCL8, but also may enable numerous negative regulations with the inhibition of EGFR [20] thus minimising the proliferative signalling [21].
Other proteins modulate the EGFR, either hindering internalisation, such as annexin A6 [22], or inducing endosomal recycling, such as the p53 protein when overexpressed, or the DRAM1 [23] and NSD2 proteins that exacerbate signalling [24], or with the Tinagl1 protein that suppresses several signalling pathways [25], modifying the action of anti-EGFR [9]. Oestrogen signalisation with EGFR results in the synthesis of the SDF-1 (CXCL12) and its receptor CXCR4, thus activating HER2, triggered by the family of Src kinases [26]. And Src kinases stimulate STAT3s which, again, intensify the modulation of EGFR [27], through intracellular signalling [28].
Overexpression of EGFR in tumour cells has been considered a poor prognostic factor in breast cancer for decades [29–31], with rare disagreements [32]. EGFR overexpression in breast tumours can be high, varying in intensity in different tumour subtypes [33, 34]. In TN neoplasms, EGFR overexpression in the tumour is found in 15%–30% of cases, with a worse prognosis especially in the presence of intense nuclear expression of EGFR [15].
The receptors of neoplastic cells can be measured in serum, through their soluble form as soluble HER-2 (sHER2), reflecting the expression of HER2 in the membrane [35]. The EGFR measured in tumour cells by IH or FISH [36] can also be measured in plasma or serum [37] and is routinely detected in patients with lung cancer [38]. The detection of EGFR as a protein expressed in the serum of breast cancer patients may be a non-invasive practice with better prognostic stratification.
This study aims to analyse the serological profile of soluble EGFR (sEGFR) in different breast cancer subgroups and its possible value as a prognostic marker, and to verify the relationship of sEGFR with clinicopathological variables and with the EGFR present in the tumour cell [tumour EGFR (tEGFR)].
Methodology
A prospective study enrolling newly diagnosed breast cancer patients treated between 2015 and 2019 was conducted at the Oncology Sector of the Clinical Hospital of the Federal University of Uberlandia, Brazil, after approval by the Ethics Committee for Research on Humans (294,508/13). The healthy control group consisted of volunteers from this hospital. All participants read and signed the Informed Consent Form. This study did not interfere with the patients’ treatment.
The sample was categorised into four groups. Three groups of patients had a biopsy showing histologically infiltrating carcinoma of the breast, but had not yet undergone surgery or treatment with chemotherapy, radiotherapy and/or hormone therapy. Staging followed the American Joint Committee on Cancer Seventh Edition [39], grouping patients into early tumours (stages I/II), locally advanced tumours (stages III) and metastatic tumours (stages IV). The fourth group consisted of healthy volunteers, with BIRADS I and II mammograms performed within the last 12 months. A questionnaire was applied to obtain information on clinical aspects, family history, use of oral contraceptives and other drugs, hormone replacement therapies and smoking [40].
Exclusion criteria for patients were second neoplasm or another severe systemic disease, such as rheumatologic, cardiac, renal or hepatic, diagnosed at inclusion or during the study. For the control group, the exclusion criterion was the diagnosis of any neoplasm or serious systemic diseases mentioned above.
Peripheral blood samples were collected from the control group and, before any treatment of the patients, placed in collection tubes in separator gel and activator to accelerate coagulation. The sEGFR measurement was performed using the sandwich Enzyme-Linked Immunosorbent Assay (ELISA) kit #DY231 (R&D Systems) according to the manufacturer’s recommendations. The reading was performed in a GloMax Explorer (Promega). The sEGFR concentrations were determined by the equation obtained from the standard curve concentrations.
In addition, the paraffin blocks were submitted to histological sections of tumor lesions, stained with hematoxylin-eosin, selecting areas of greater neoplastic representation, and mounted for optimized analysis in tissue samples array (tissue microarray (TMA)) [41] of according to the routine of a high-demand pathology laboratory [42]. The antibody employed to detect tEGFR was Dako’s clone 31G7, analysed by two pathologists at different times. When the two disagreed in their analysis, a discussion was established until consensus was reached. In the absence of any reaction/staining, the result was negative. The presence of any reaction/staining was considered positive, ranging from 1% to 90%, stratified as follows: <10% weak positive; ≥10% and <40% intermediate positive; and ≥40% strong positive.
Statistical analysis
Data were analysed using the software packages IBM SPSS 25.0, Jamovi 1.6.7 and GraphPad Prism 8.0. Statistical significance was defined as p < 0.05.
The Shapiro–Wilk statistical test verified data distribution, and descriptive data were represented according to the obtained distributions. Student’s t-test or Mann–Whitney U test was utilised to assess the difference between the two groups. ANOVA or Kruskal–Wallis was utilised to test the difference between more than two groups, with Tukey or Dunn’s post hoc, respectively.
To assess bivariate correlation in the presence of an ordinal or categorical variable, or in the presence of a continuous, non-symmetric/normal variable, Spearman’s correlation test was used. The association between interdependent categorical factors was evaluated using Pearson’s χ2 test. The association was considered positive (direct) when the adjusted standardised residuals had a value >(+2.0) and considered negative (indirect/inverse) when the value was <(–2.0).
The survival curve with continuous variables was used for determining the optimal cutoff points and prognostic factors of continuous variables. For survival analysis of categorical variables, the Kaplan–Meier estimator was employed to evaluate the proportionality of risks as a requirement for Cox regression.
Results
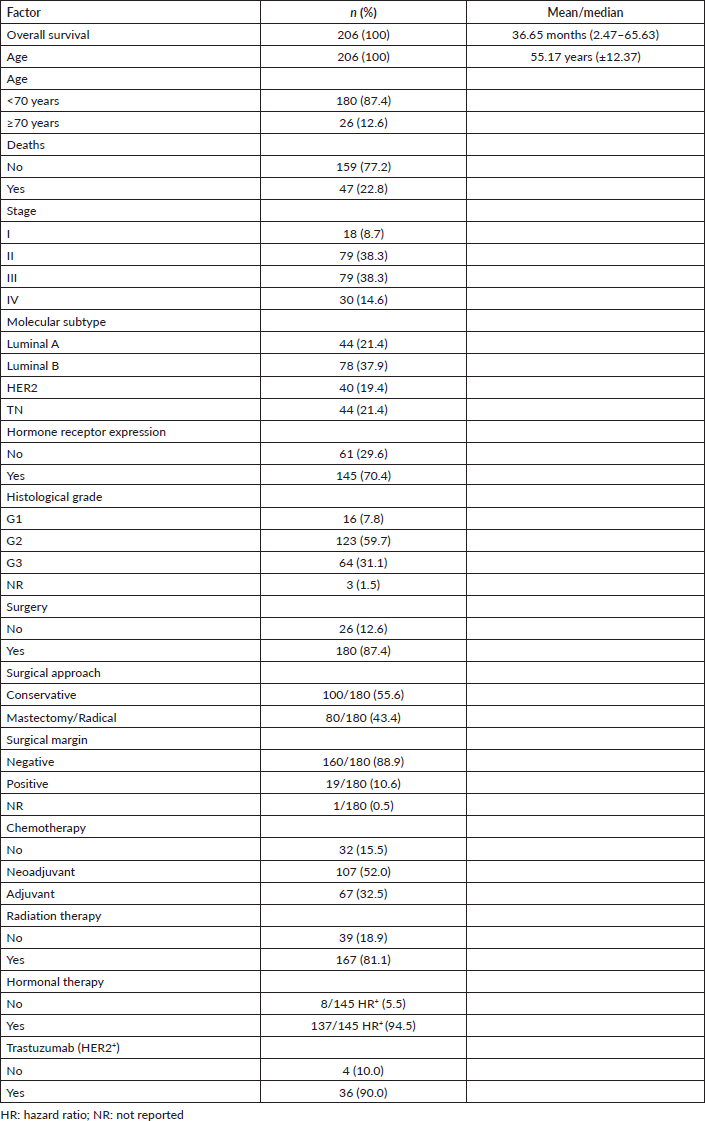
This study included 206 patients with breast carcinoma and 89 healthy age-matched women. In a median follow-up period of 36.6 months, 47 deaths occurred. Twenty-six patients were not operated on, 20 for being metastatic and 6 due to early death, high surgical risk or progression during neoadjuvant chemotherapy. The results are reported according to the guidelines established by Strengthening the Reporting of Observational Studies in Epidemiology. Other clinical and therapeutic characteristics are shown in Table 1.
The sEGFR level from patients (30.70 ng/mL, range: 4.78–100.85) differed significantly (Mann–Whitney U p < 0.0005) from healthy controls (40.45 ng/mL, range: 18.10–117.15) (Figure 1a). Significantly greater sEGFR levels were observed in healthy controls compared to all breast cancer subtypes (Kruskal–Wallis p < 0.0005; Dunn’s p at least < 0.05 in all pairwise comparisons), but no difference was observed among molecular subtypes (Dunn’s p > 0.999 for all pairwise comparisons) (Figure 1b).
Table 1. Clinical, laboratory and treatment characteristics of patients (n = 206).
Subsequently, survival analyses were performed to test whether sEGFR has any prognostic value. In accordance with the exploratory nature of this study, survival analyses for continuous variables were performed for the entire cohort as well as subgroup analyses to obtain the best sEGFR cutoff predictive for survival in each context.
In the analysis of the sEGFR concentration among the 206 patients, although not significant (Log-Rank p = 0.073), a shorter overall survival (OS) was observed in those with the highest concentration of sEGFR (Figure 2a). Exclusion of the 44 TN patients resulted in a high increase of the p-value (Log-Rank p = 0.389) (Figure 2b).
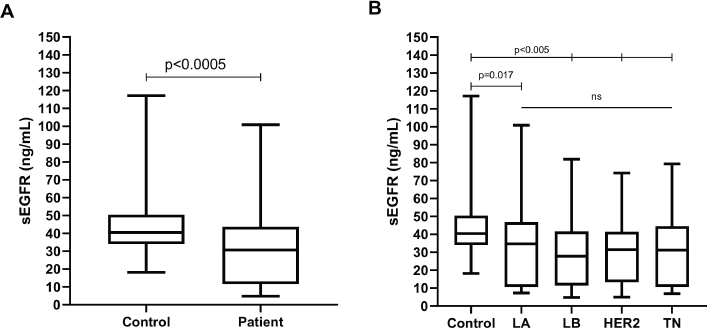
Figure 1. Boxplot representing the peripheral blood serum EGFR (sEGFR) levels (ng/mL) in (a): healthy volunteers (n = 89) versus breast cancer patients (n = 206) and (b): healthy volunteers versus breast cancer patients (n = 206), according to IH subtype (LA, Luminal A; LB, Luminal B; HER2, HER2/ERBB2 overpexression/amplification; TN, triple-negative).
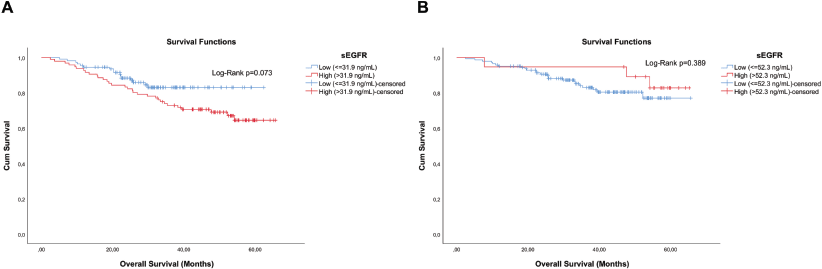
Figure 2. Cumulative survival curves by the KM estimator according to sEGFR levels. (a): Overall survival in the entire cohort (n = 206) and (b): non-TN breast cancer patients (n = 162). Each group was submitted to analysis for establishing the optimal sEGFR level cutoff with prognostic value. Lower sEGFR levels were marginally associated with increased overall survival (blue curve), compared to those with higher levels (red curve), but only in the entire cohort including TN breast cancer patients.
The 96 patients with sEGFR > 31.9 ng/mL had a median OS of 30.27 months (range: 5.07–62.40), significantly lower (Mann–Whitney U p < 0.0005) than the 110 patients with sEGFR ≤ 31.9 ng/mL (median OS of 46.55 months (range: 2.47–65.63)). Again, when TN patients are excluded from these analyses, this difference in OS disappears (Mann–Whitney U p = 0.728). In the OS analysis, the sEGFR levels, alongside the immunohistochemistry type and stage, were associated with prognosis (Table 2). A higher risk of death was observed in patients with Luminal B tumours, and more intensely with TN tumours, compared to patients with lower risk such as Luminal A tumours, and also with patients with HER2+ tumours but using trastuzumab. The type of surgery did not interfere with OS (p = 0.324).
Analysing the expression of sEGFR in the IH subtypes, we found that in the subgroups of patients with Luminal A, Luminal B and HER2+ tumours, there was a non-significant (p > 0.05) difference in OS.
TN breast cancer patients (n = 44) with sEGFR ≤ 47.8 ng/mL (37 patients) had a higher median OS (27.33 months (range: 6.63–62.40)) than those with sEGFR > 47.8 ng/mL (13.37 months (range: 3.67–60.43)) (Figure 3). By multivariable Cox regression, the high sEGFR (>47.8 ng/mL) experienced a death risk increase of more than 400% during the follow-up (adjusted HR: 5.149 (1.900–13.955), p = 0.001) (Table 3). The subgroup with low sEGFR expression, with longer survival between TN tumours, had lower OS than patients with Luminal A (p = 0.0004) and HER2+ (p = 0.0130) tumours, but with no OS difference in relation to patients with luminal B tumours (p = 0.0991).
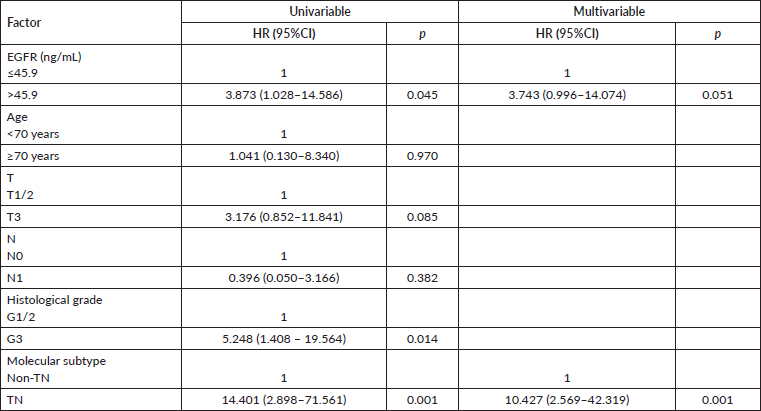
Our analysis of patients with early tumours (I+II; n = 97) revealed that the TN subtype showed significantly reduced OS in univariable and multivariable analyses (Table 4). Patients with early tumours with sEGFR expression > 45.9 ng/mL presented, in univariable analysis, higher OS than those with lower sEGFR values (p = 0.045), but without significance in multivariable analysis (p = 0.051). And, again, when TN patients were excluded from these initial tumours, there was no significant difference in the analyses (univariable Cox regression p = 0.609). In patients in stages III (79 patients; univariable Cox regression p = 0.438) or IV (30 patients; univariable Cox regression p = 0.965), the OS was not influenced by high or low sEGFR expression.
There was no significant correlation of sEGFR with histological tumour grade (G1/G2/G3; Spearman’s rho of −0.003; p = 0.972), Ki67 (Spearman’s rho of −0.004; p = 0.956), BMI, tumour size (Spearman’s rho of −0.011; p = 0.876), lymph node metastasis (Spearman’s rho of −0.090; p = 0.225), nor stage (Spearman’s rho of −0.045; p = 0.522). Similarly, there was no significant difference in the median sEGFR levels in relation to the categories of histological tumour grade (Kruskal–Wallis p = 0.996), Ki67 (< or ≥14%) (Mann–Whitney U p = 0.518), tumour size (Kruskal–Wallis p = 0.246), lymph node metastasis (Kruskal–Wallis p = 0.372) nor stage (Kruskal–Wallis p = 0.822).
The initial tested cutoff (sEGFR > 31.9 ng/mL) was also not associated with histological grade (Pearson’s χ2: 0.129, p = 0.938), high ki67 (≥14%) levels (Pearson’s χ2: 0.567, p = 0.451), tumour size (Pearson’s χ2: 5.087, p = 0.166), lymph node metastasis (Pearson’s χ2: 4.734, p = 0.192), stage (Pearson’s χ2: 1.109, p = 0.775) nor immunohistochemistry subtype (Pearson’s χ2: 2.086, p = 0.555). Similar non-significant results were observed in the analysis of exclusive TN cancers by its obtained cutoff (>47.8 ng/mL).
The expression of tEGFR was analysed in 94 patients, equally representing all stages and IH subtypes, of which 66 specimens were negative and 28 (29.78%) were positive in ≥ 1% in tumour cells, with 11 weak positives, 6 intermediate positives and 11 strong positives. There was no correlation between tEGFR and sEGFR (Spearman’s rho of −0.022; p = 0.852). Negative or positive tEGFR had no impact on OS (Log-Rank p = 0.518). There was an inverse correlation between tEGFR and Oestrogen Receptor (ER) (Spearman’s rho of −0.752; p < 0.005). ER-negative breast cancers expressed more tEGFR (Mann–Whitney U p < 0.0001).
Discussion
The IH subtypes and stages directly impact breast cancer prognosis. Compared to Luminal A and HER2+ (when properly treated with trastuzumab) subtypes, Luminal B and TN subtypes have shorter survivals. Also, locally advanced and metastatic tumours (stages III and IV) have significantly shorter OS than initial-stage ones [43–45]. In the current study, the survival of patients was similar to other treatment centres, varying by prognostic characteristics and geographic regions [46]. Patients with more aggressive HER2+ tumours receiving anti-HER2 therapy had more prolonged survival than those with Luminal B tumours. On the other hand, predictive factors and an effective target therapy remain lacking in patients with TN breast tumours, who continue to face poorer survival.
Table 2. Univariable and multivariable analyses of overall survival of breast cancer patients (n = 206).
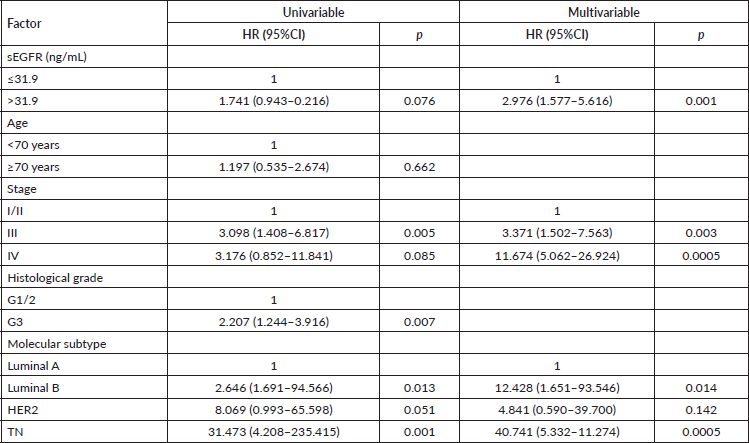
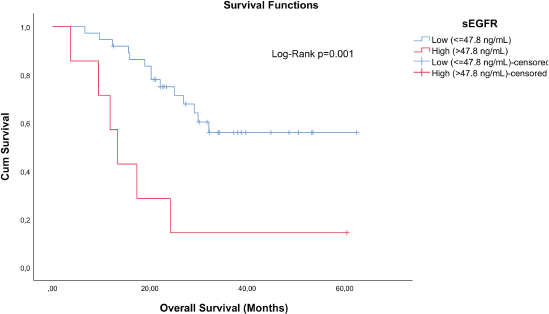
Figure 3. Cumulative survival curves by the KM estimator according to sEGFR levels in TN patients (n = 44). Lower sEGFR levels were associated with increased overall survival (blue curve), compared to those with higher levels (red curve).
Table 3. Multivariable analysis of overall survival of patients with TN breast tumour (n = 44).
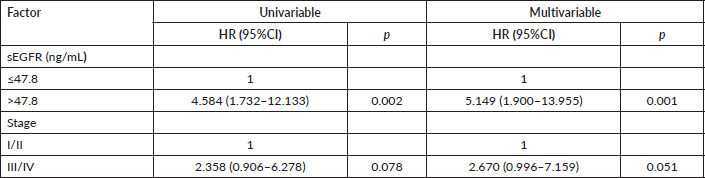
Table 4. Univariable and multivariable analyses of overall survival in subgroups of patients with early (stages I + II) breast tumours (n = 97).
Only 94 paraffin blocks of tumours were analysed, but with representation proportional to the stages and IH subtypes. The technique for analysing tEGFR by TMA is reproducible like the individualised IH [47]. Our tEGFR analysis was hampered by several technical issues. We planned to analyse the expression of EGFR in the tumour in at least 50% of the cases under study, already foreseeing the difficulty of accessing this material since many patients undergo biopsy or undergo surgery in other services and then are referred to our centre for treatment. Of the 116 paraffin blocks accessed in 22 samples, there was no viable tissue, resulting from fine needle biopsies or damaged by non-ideal storage [48]. The expression of tEGFR did not interfere with OS, perhaps because of our small sample number. A similar study [49] but with a larger number of patients showed that tEGFR overexpressed in TN tumors, detected by immunohistochemistry [30], also worsened survival. We also did not specify the location of tEGFR, as the nuclear expression has a worse prognosis than tEGFR expression in the cytoplasmic membrane [50]. We demonstrated a significant inverse correlation between tEGFR and ER expression, although without specifying whether the tumour cells were ERβ or ERα, which have different expressions of tEGFR [51]. Despite these deficiencies, we also did not find a correlation between tEGFR and sEGFR, similar to other studies [52, 53]. Due to the difficulties presented – such as scarcity of material, difficulty in ideal storage and the possibility of constant recycling of tEGFR – the measurement of sEGFR by the ELISA method may allow the analysis of this tumour aggressiveness parameter, with greater precision.
The sEGFR concentration in all patients was significantly lower than in healthy women, a result that is conflicting in the literature and lacks a rational explanation [54–56]. But perhaps in healthy women, there is a necessity for positive regulations for growth and physiological division, without the numerous possible negative regulations, such as the internalisation of these overexpressed receptors and other EGFR inhibitions in an attempt to minimise the proliferative signalling of the neoplasia. Therefore, sEGFR is not a biomarker for population screening. The sEGFR measurements in cancer patients are also contradictory but can be explained by the methodological disparities among the studies.
Analyses of women with metastatic breast cancer have reported low levels of sEGFR, but with normal sHER2, being related to shorter survival [57]; or high concentrations associated with longer survival [54]; or absence in the survival correlation with the sEGFR [52, 53]. In early breast tumours, there is a study correlating low sEGFR expression with lower survival [58]. We did not find this difference in the survival of patients with early or advanced tumours.
We suggest that sEGFR is not a promising biomarker to stratify higher or lower risk patients among those with luminal A, B or HER2+ tumours. However, in the subgroup with TN tumours, the sEGFR distinguished in early and advanced tumours, with statistical significance, two risk groups. In the low expression of sEGFR, there was longer survival, but significantly lower than the OS of Luminal A or HER2+ patients and equal to the survival of patients with Luminal B tumours. This TN group with low sEGFR expression would be the group with the least poor prognosis. And a second TN group with an even worse prognosis is those patients with high sEGFR expression and a four-fold greater death risk. In EGFR gene signalling, there is overexpression of other proteins, and when there is simultaneous signalling of the HER2 gene, the Snail [59] protein is overexpressed, with an evident deterioration in survival. There are improvements in this prognosis by pharmacological modulation, such as the efficient use of anti-HER2 therapy, as probably occurred in the patients in the current study who expressed EGFR and HER2. We did not measure the sHER2 protein, but together with the sEGFR, perhaps the treatment response can be measured in patients with metastatic breast cancer [60].
Despite progress in the treatment of women with TN breast tumours, there is a need to stratify among TN tumours those with better or worse prognosis, de-escalating treatment in women with less aggressive tumours. We need both greater individualisation and more predictive markers for the proper treatment of TN tumours. Perhaps the use of PARP inhibitors in mutated BRCA1/2 should be consolidated in the adjuvant setting [61], or when the ideal association is found for taxane and anthracycline such as carboplatin, pembrolizumab or atezolizumab in neoadjuvant treatment [62]. And in the metastatic setting, the benefit of immunotherapy associated with chemotherapy has already been demonstrated [63, 64] with promising new formulations such as sacituzumab govitecan-hziy [65]. And research continues targeting EGFR in TN tumours. The use of cetuximab was disappointing, due to its toxicity [66] and few objective responses [67]. But the combination of cetuximab with panitumumab proved to be effective in EGFR degradation and tumour reduction in an experimental study [68]. And new presentations of cetuximab, in in vitro studies, linked to nanoparticles minimised treatment resistance [69], through promising associations with bioconjugated molecules [70, 71], or with titanium, which hinders the action of ligands on EGFR [72].
Another possible inhibition of EGFR is through the use of TKIs. Preclinical studies have shown that gefitinib increased the response to carboplatin and docetaxel [73]; doxazosin reduced EGFR expression [74]; epertinib inhibited EGFR and HER2 [75]; almonertinib inhibited the multi-resistance of cancer cells [76]. The association of erlotinib and metformin minimised signalling [77] and the inhibition of EGFR and PI3K increased the sensitivity to irradiation [78]. Everolimus associated with gefitinib inhibited the PI3K/AKT/mTOR pathways [79], and gefitinib with anastrozole showed an objective [80] but irregular response to TKI due to the heterogeneity of EGFR [81]. A small clinical study in patients with advanced bowel cancer, having sEGFR measured, used FOLFOX6 and gefitinib, demonstrating a more objective response in those with higher sEGFR [82]. Research studies directed to the management of EGFR [83], including new gene therapies [84], demonstrated the feasibility of the suppression of the Notch3 gene with reduced internalisation of EGFR [85], or the construction of mutant forms of the EGF ligand blocking this receptor [86], or the genetic silencing by small RNAs of interference (siRNA) against EGFR [87]. New therapies, new antibody formations or new associations with TKIs signal the importance of adequately measuring EGFR, both as a prognostic biomarker, and as a treatment predictor, in women with TN breast tumours.
This study has several limitations. Despite its prospective nature and assessment of patients from the year 2015 onwards, most of whom received adequate treatments according to current guidelines, the short follow-up period and low sample size make the external validity very low, thus requiring caution when extrapolating these data to any and all patients with breast cancer, especially the TN subtype. Therefore, the results must be understood as exploratory, requiring multicentre data and the follow-up of a greater number of patients for a longer period of time.
Conclusion
EGFR measurement is inexpensive and straightforward and can be measured in TN breast cancer. Increased sEGFR expression significantly increased the death risk in TN tumours, but not in Luminals or HER2+. Future studies should confirm our findings and allow different therapeutic options depending on the serum concentration of this biomarker, reconfiguring two large groups of TN tumours: one with a less-bad prognosis and another TN group with an even worse prognosis.
Acknowledgments
The authors thank the volunteer women of this study.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Funding statement
This work was supported by Grupo Luta Pela Vida. The authors received no financial support for authorship and/or publication of this article.
Data
Data is deposited in Mendeley Data (DOI: 10.17632/xxt6mc2pk7.3)
References
1. Sung H, Ferlay J, and Siegel RL, et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 71(3) 209–249 https://doi.org/10.3322/caac.21660 PMID: 33538338
2. Kerlikowske K, Grady D, and Barclay J, et al (1996) Effect of age, breast density, and family history on the sensitivity of first screening mammography JAMA 276(1) 33–38 https://doi.org/10.1001/jama.1996.03540010035027 PMID: 8667536
3. Warner E, Plewes DB, and Hill KA, et al (2004) Surveillance of BRCA1 and BRCA2 mutation carriers with magnetic resonance imaging, ultrasound, mammography, and clinical breast examination JAMA 292(11) 1317–1325 https://doi.org/10.1001/jama.292.11.1317 PMID: 15367553
4. Lord SJ, Lei W, and Craft P, et al (2007) A systematic review of the effectiveness of magnetic resonance imaging (MRI) as an addition to mammography and ultrasound in screening young women at high risk of breast cancer Eur J Cancer 43(13) 1905–1917 https://doi.org/10.1016/j.ejca.2007.06.007 PMID: 17681781
5. Giuliano AE, Connolly JL, and Edge SB, et al (2017) Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual CA Cancer J Clin 67(4) 290–303 https://doi.org/10.3322/caac.21393 PMID: 28294295
6. Slamon DJ, Godolphin W, and Jones LA, et al (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer Science 244(4905) 707–712 https://doi.org/10.1126/science.2470152 PMID: 2470152
7. Sparano JA, Crager MR, and Tang G, et al (2021) Development and validation of a tool integrating the 21-gene recurrence score and clinical-pathological features to individualize prognosis and prediction of chemotherapy benefit in early breast cancer J Clin Oncol 39(6) 557–564 https://doi.org/10.1200/JCO.20.03007 PMCID: 8078482
8. Denduluri N, Somerfield MR, and Chavez-MacGregor M, et al (2021) Selection of optimal adjuvant chemotherapy and targeted therapy for early breast cancer: ASCO guideline update J Clin Oncol 39(6) 685–693 https://doi.org/10.1200/JCO.20.02510
9. Shapira I, Lee A, and Vora R, et al (2013) P53 mutations in triple negative breast cancer upregulate endosomal recycling of epidermal growth factor receptor (EGFR) increasing its oncogenic potency Crit Rev Oncol Hematol 88(2) 284–292 https://doi.org/10.1016/j.critrevonc.2013.05.003 PMID: 23755891
10. Byeon HK, Ku M, and Yang J (2019) Beyond EGFR inhibition: multilateral combat strategies to stop the progression of head and neck cancer Exp Mol Med 51(1) 1–14 https://doi.org/10.1038/s12276-018-0202-2 PMID: 30700700 PMCID: 6353966
11. Gonzalez-Conchas GA, Rodriguez-Romo L, and Hernandez-Barajas D, et al (2018) Epidermal growth factor receptor overexpression and outcomes in early breast cancer: a systematic review and a meta-analysis Cancer Treat Rev 62 1–8 https://doi.org/10.1016/j.ctrv.2017.10.008
12. Constantinou C, Papadopoulos S, and Karyda E, et al (2018) Expression and clinical significance of Claudin-7, PDL-1, PTEN, c-Kit, c-Met, c-Myc, ALK, CK5/6, CK17, p53, EGFR, Ki67, p63 in triple-negative breast cancer-a single centre prospective observational study In Vivo 32(2) 303–311 PMID: 29475913 PMCID: 5905198
13. Rao C, Shetty J, and Prasad KH (2013) Immunohistochemical profile and morphology in triple - negative breast cancers J Clin Diagn Res 7(7) 1361–1365 PMID: 23998066 PMCID: 3749636
14. Colomiere M, Ward AC, and Riley C, et al (2009) Cross talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas Br J Cancer 100(1) 134–144 https://doi.org/10.1038/sj.bjc.6604794 PMCID: 2634691
15. Hsu JL and Hung MC (2016) The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer Cancer Metastasis Rev 35(4) 575–588 https://doi.org/10.1007/s10555-016-9649-6 PMID: 27913999 PMCID: 5215954
16. Papanastasiou AD, Sirinian C, and Plakoula E, et al (2017) RANK and EGFR in invasive breast carcinoma Cancer Genet 216–217 61–66 https://doi.org/10.1016/j.cancergen.2017.07.004
17. Capuani F, Conte A, and Argenzio E, et al (2015) Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells Nat Commun 6 7999 https://doi.org/10.1038/ncomms8999 PMID: 26264748 PMCID: 4538861
18. Zanetti-Domingues LC, Bonner SE, and Martin-Fernandez ML, et al (2020) Mechanisms of action of EGFR tyrosine kinase receptor incorporated in extracellular vesicles Cells 9(11) 2505 https://doi.org/10.3390/cells9112505 PMCID: 7699420
19. Guo G, Gong K, and Wohlfeld B, et al (2015) Ligand-independent EGFR signaling Cancer Res 75(17) 3436–3441 https://doi.org/10.1158/0008-5472.CAN-15-0989 PMID: 26282175 PMCID: 4558210
20. Suenaga M, Stintzing S, and Cao S, et al (2019) Role of CCL5 and CCR5 gene polymorphisms in epidermal growth factor receptor signalling blockade in metastatic colorectal cancer: analysis of the FIRE-3 trial Eur J Cancer 107 100–114 https://doi.org/10.1016/j.ejca.2018.11.019
21. Ahmed S, Mohamed HT, and El-Husseiny N, et al (2021) IL-8 secreted by tumor associated macrophages contribute to lapatinib resistance in HER2-positive locally advanced breast cancer via activation of Src/STAT3/ERK1/2-mediated EGFR signaling Biochim Biophys Acta Mol Cell Res 1868(6) 118995 https://doi.org/10.1016/j.bbamcr.2021.118995 PMID: 33667527
22. Korolkova OY, Widatalla SE, and Williams SD, et al (2020) Diverse roles of annexin A6 in triple-negative breast cancer diagnosis, prognosis and EGFR-targeted therapies Cells 9(8) 1855 https://doi.org/10.3390/cells9081855 PMCID: 7465958
23. Geng J, Zhang R, and Yuan X, et al (2020) DRAM1 plays a tumor suppressor role in NSCLC cells by promoting lysosomal degradation of EGFR Cell Death Dis 11(9) 768 https://doi.org/10.1038/s41419-020-02979-9 PMID: 32943616 PMCID: 7498585
24. Wang JJ, Zou JX, and Wang H, et al (2019) Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling Acta Pharmacol Sin 40(8) 1067–1075 https://doi.org/10.1038/s41401-018-0199-z PMID: 30670815 PMCID: 6786427
25. Shen M, Jiang YZ, and Wei Y, et al (2019) Tinagl1 suppresses triple-negative breast cancer progression and metastasis by simultaneously inhibiting integrin/FAK and EGFR signaling Cancer Cell 35(1) 64–80 e7 https://doi.org/10.1016/j.ccell.2018.11.016 PMID: 30612941
26. Almendro V, Garcia-Recio S, and Gascon P (2010) Tyrosine kinase receptor transactivation associated to G protein-coupled receptors Curr Drug Targets 11(9) 1169–1180 https://doi.org/10.2174/138945010792006807 PMID: 20450475
27. Balanis N and Carlin CR (2017) Stress-induced EGF receptor signaling through STAT3 and tumor progression in triple-negative breast cancer Mol Cell Endocrinol 451 24–30 https://doi.org/10.1016/j.mce.2017.01.013 PMID: 28088463 PMCID: 5469704
28. Medts T, de Diesbach P, and Cominelli A, et al (2010) Acute ligand-independent Src activation mimics low EGF-induced EGFR surface signalling and redistribution into recycling endosomes Exp Cell Res 316(19) 3239–3253 https://doi.org/10.1016/j.yexcr.2010.09.001 PMID: 20832399
29. Sainsbury JR, Farndon JR, and Needham GK, et al (1987) Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer Lancet 1(8547) 1398–1402 PMID: 2884496
30. Abdelrahman AE, Rashed HE, and Abdelgawad M, et al (2017) Prognostic impact of EGFR and cytokeratin 5/6 immunohistochemical expression in triple-negative breast cancer Ann Diagn Pathol 28 43–53 https://doi.org/10.1016/j.anndiagpath.2017.01.009 PMID: 28648939
31. Nieto Y, Nawaz F, and Jones RB, et al (2007) Prognostic significance of overexpression and phosphorylation of epidermal growth factor receptor (EGFR) and the presence of truncated EGFRvIII in locoregionally advanced breast cancer J Clin Oncol 25(28) 4405–4413 https://doi.org/10.1200/JCO.2006.09.8822 PMID: 17906204
32. Ferrero JM, Ramaioli A, and Largillier R, et al (2001) Epidermal growth factor receptor expression in 780 breast cancer patients: a reappraisal of the prognostic value based on an eight-year median follow-up Ann Oncol 12(6) 841–846 https://doi.org/10.1023/A:1011183421477 PMID: 11484962
33. Opstal-van Winden AW, Krop EJ, and Karedal MH, et al (2011) Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study BMC Cancer 11 381 https://doi.org/10.1186/1471-2407-11-381 PMID: 21871081 PMCID: 3189190
34. Li CI (2011) Discovery and validation of breast cancer early detection biomarkers in preclinical samples Horm Cancer 2(2) 125–131 https://doi.org/10.1007/s12672-010-0061-3 PMID: 21761335 PMCID: 3228358
35. Fehm T, Jager W, and Kramer S, et al (2004) Prognostic significance of serum HER2 and CA 15-3 at the time of diagnosis of metastatic breast cancer Anticancer Res 24(3b) 1987–1992 PMID: 15274389
36. Gumuskaya B, Alper M, and Hucumenoglu S, et al (2010) EGFR expression and gene copy number in triple-negative breast carcinoma Cancer Genet Cytogenet 203(2) 222–229 https://doi.org/10.1016/j.cancergencyto.2010.07.118 PMID: 21156237
37. Wu Y, Liu H, and Shi X, et al (2015) Can EGFR mutations in plasma or serum be predictive markers of non-small-cell lung cancer? A meta-analysis Lung Cancer 88(3) 246–253 https://doi.org/10.1016/j.lungcan.2015.03.008 PMID: 25837799
38. Hiley CT, Le Quesne J, and Santis G, et al (2016) Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease Lancet 388(10048) 1002–1011 https://doi.org/10.1016/S0140-6736(16)31340-X PMID: 27598680
39. Edge SB, Byrd DR, and Compton CC, et al (2015) AJCC Cancer Staging Manual 7th edn (New York: Springer-Verlag)
40. Pitteri SJ and Hanash SM (2011) Confounding effects of hormone replacement therapy in protein biomarker studies Cancer Epidemiol Biomarkers Prev 20(1) 134–139 https://doi.org/10.1158/1055-9965.EPI-10-0673
41. Koo M, Squires JM, and Ying D, et al (2019) Making a tissue microarray Methods Mol Biol 1897 313–323 https://doi.org/10.1007/978-1-4939-8935-5_27
42. O Leary PC, Penny SA, and Dolan RT, et al (2013) Systematic antibody generation and validation via tissue microarray technology leading to identification of a novel protein prognostic panel in breast cancer BMC Cancer 13 175 https://doi.org/10.1186/1471-2407-13-175 PMID: 23547718 PMCID: 3668187
43. de Araujo RA, Cordero da Luz FA, and da Costa Marinho E, et al (2021) Operable breast cancer: how not to worsen the prognosis, especially in triple negative and stage II tumors Surg Oncol 38 101596 https://doi.org/10.1016/j.suronc.2021.101596 PMID: 34015750
44. da Luz FAC, da Costa Marinho E, and Nascimento CP, et al (2022) The benefits of trastuzumab in the treatment of HER2+ breast cancer as a function of exposure time Ecancermedicalscience 16 1347 PMID: 35242228 PMCID: 8831111
45. Luz F, Marinho EDC, and Nascimento CP, et al (2022) The effectiveness of radiotherapy in preventing disease recurrence after breast cancer surgery Surg Oncol 41 101709 https://doi.org/10.1016/j.suronc.2022.101709 PMID: 35124329
46. Oliveira NPD, Cancela MC, and Martins LFL, et al (2021) Spatial distribution of advanced stage diagnosis and mortality of breast cancer: socioeconomic and health service offer inequalities in Brazil PLoS One 16(2) e0246333 https://doi.org/10.1371/journal.pone.0246333 PMID: 33534799 PMCID: 7857585
47. Dekker TJ, ter Borg S, and Hooijer GK, et al (2015) Quality assessment of estrogen receptor and progesterone receptor testing in breast cancer using a tissue microarray-based approach Breast Cancer Res Treat 152(2) 247–252 https://doi.org/10.1007/s10549-015-3444-x PMID: 26041687 PMCID: 4491103
48. Omilian AR, Zirpoli GR, and Cheng TD, et al (2020) Storage conditions and immunoreactivity of breast cancer subtyping markers in tissue microarray sections Appl Immunohistochem Mol Morphol 28(4) 267–273 https://doi.org/10.1097/PAI.0000000000000756
49. Liu D, He J, and Yuan Z, et al (2012) EGFR expression correlates with decreased disease-free survival in triple-negative breast cancer: a retrospective analysis based on a tissue microarray Med Oncol 29(2) 401–405 https://doi.org/10.1007/s12032-011-9827-x
50. Lo HW (2010) Nuclear mode of the EGFR signaling network: biology, prognostic value, and therapeutic implications Discov Med 10(50) 44–51 PMID: 20670598 PMCID: 3637667
51. Skandalis SS, Afratis N, and Smirlaki G, et al (2014) Cross-talk between estradiol receptor and EGFR/IGF-IR signaling pathways in estrogen-responsive breast cancers: focus on the role and impact of proteoglycans Matrix Biol 35 182–193 https://doi.org/10.1016/j.matbio.2013.09.002
52. Witzel I, Thomssen C, and Krenkel S, et al (2006) Clinical utility of determination of HER-2/neu and EGFR fragments in serum of patients with metastatic breast cancer Int J Biol Markers 21(3) 131–140 https://doi.org/10.1177/172460080602100301 PMID: 17013794
53. Hudelist G, Kostler WJ, and Gschwantler-Kaulich D, et al (2006) Serum EGFR levels and efficacy of trastuzumab-based therapy in patients with metastatic breast cancer Eur J Cancer 42(2) 186–192 https://doi.org/10.1016/j.ejca.2005.08.036
54. Banys-Paluchowski M, Witzel I, and Riethdorf S, et al (2017) Evaluation of serum epidermal growth factor receptor (EGFR) in correlation to circulating tumor cells in patients with metastatic breast cancer Sci Rep 7(1) 17307 https://doi.org/10.1038/s41598-017-17514-8 PMID: 29229933 PMCID: 5725555
55. Kjaer IM, Olsen DA, Brandslund I, et al (2020) Dysregulated EGFR pathway in serum in early-stage breast cancer patients: a case control study Sci Rep 10(1) 6714 https://doi.org/10.1038/s41598-020-63375-z PMID: 32317675 PMCID: 7174424
56. Tas F, Bilgin E, and Karabulut S, et al (2015) Clinical significance of serum epidermal growth factor receptor (EGFR) levels in patients with breast cancer Cytokine 71(1) 66–70 https://doi.org/10.1016/j.cyto.2014.09.001
57. Souder C, Leitzel K, and Ali SM, et al (2006) Serum epidermal growth factor receptor/HER-2 predicts poor survival in patients with metastatic breast cancer Cancer 107(10) 2337–2345 https://doi.org/10.1002/cncr.22255 PMID: 17048231
58. Kjaer IM, Olsen DA, and Brandslund I, et al (2020) Prognostic impact of serum levels of EGFR and EGFR ligands in early-stage breast cancer Sci Rep 10(1) 16558 https://doi.org/10.1038/s41598-020-72944-1 PMID: 33024132 PMCID: 7538553
59. Chang HY, Tseng YK, Chen YC, et al High snail expression predicts a poor prognosis in breast invasive ductal carcinoma patients with HER2/EGFR-positive subtypes Surg Oncol 27(2) 314–320 PMID: 29937187
60. Sandri MT, Johansson HA, and Zorzino L, et al (2007) Serum EGFR and serum HER-2/neu are useful predictive and prognostic markers in metastatic breast cancer patients treated with metronomic chemotherapy Cancer 110(3) 509–517 https://doi.org/10.1002/cncr.22825 PMID: 17559147
61. Thomssen C, Balic M, and Harbeck N, et al (2021) St. Gallen/Vienna 2021: a brief summary of the consensus discussion on customizing therapies for women with early breast cancer Breast Care (Basel) 16(2) 135–143 https://doi.org/10.1159/000516114
62. Korde LA, Somerfield MR, and Carey LA, et al (2021) Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline J Clin Oncol 39(13) 1485–1505 https://doi.org/10.1200/JCO.20.03399 PMID: 33507815 PMCID: 8274745
63. Schmid P, Adams S, and Rugo HS, et al (2018) Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer N Engl J Med 379(22) 2108–2121 https://doi.org/10.1056/NEJMoa1809615 PMID: 30345906
64. Cortes J, Cescon DW, and Rugo HS, et al (2020) Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial Lancet 396(10265) 1817–1828 https://doi.org/10.1016/S0140-6736(20)32531-9 PMID: 33278935
65. Smith SM, Wachter K, and Burris HA, 3rd, et al (2021) Clinical cancer advances 2021: ASCO’s report on progress against cancer J Clin Oncol 39(10) 1165–1184 https://doi.org/10.1200/JCO.20.03420 PMID: 33527845
66. Modi S, D’Andrea G, and Norton L, et al (2006) A phase I study of cetuximab/paclitaxel in patients with advanced-stage breast cancer Clin Breast Cancer 7(3) 270–277 https://doi.org/10.3816/CBC.2006.n.040 PMID: 16942645
67. Nechushtan H, Vainer G, and Stainberg H, et al (2014) A phase 1/2 of a combination of cetuximab and taxane for “triple negative” breast cancer patients Breast 23(4) 435–438 https://doi.org/10.1016/j.breast.2014.03.003 PMID: 24836394
68. Ferraro DA, Gaborit N, and Maron R, et al (2013) Inhibition of triple-negative breast cancer models by combinations of antibodies to EGFR Proc Natl Acad Sci USA 110(5) 1815–1820 https://doi.org/10.1073/pnas.1220763110 PMID: 23319610 PMCID: 3562774
69. Roncato F, Rruga F, Porcu E, et al (2018) Improvement and extension of anti-EGFR targeting in breast cancer therapy by integration with the Avidin-Nucleic-Acid-Nano-Assemblies Nat Commun 9(1) 4070 https://doi.org/10.1038/s41467-018-06602-6 PMID: 30287819 PMCID: 6172284
70. Inoue S, Patil R, and Portilla-Arias J, et al (2012) Nanobiopolymer for direct targeting and inhibition of EGFR expression in triple negative breast cancer PLoS One 7(2) e31070 https://doi.org/10.1371/journal.pone.0031070 PMID: 22355336 PMCID: 3280290
71. de Lavera I, Merkling PJ, Oliva JM, et al (2021) EGFR-targeting antitumor therapy: neuregulins or antibodies? Eur J Pharm Sci 158 105678 https://doi.org/10.1016/j.ejps.2020.105678
72. Kim H, Jeon D, Oh S, et al (2019) Titanium dioxide nanoparticles induce apoptosis by interfering with EGFR signaling in human breast cancer cells Environ Res 175 117–123 https://doi.org/10.1016/j.envres.2019.05.001 PMID: 31112848
73. Corkery B, Crown J, Clynes M, et al (2009) Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer Ann Oncol 20(5) 862–867 https://doi.org/10.1093/annonc/mdn710 PMID: 19150933
74. Hui H, Fernando MA, and Heaney AP (2008) The alpha1-adrenergic receptor antagonist doxazosin inhibits EGFR and NF-kappaB signalling to induce breast cancer cell apoptosis Eur J Cancer 44(1) 160–166 https://doi.org/10.1016/j.ejca.2007.10.002
75. Arkenau HT, Italiano A, and Mak G, et al (2018) An extended phase Ib study of epertinib, an orally active reversible dual EGFR/HER2 tyrosine kinase inhibitor, in patients with solid tumours Eur J Cancer 103 17–23 https://doi.org/10.1016/j.ejca.2018.07.134 PMID: 30196106
76. Wu CP, Hung TH, and Lusvarghi S, et al (2021) The third-generation EGFR inhibitor almonertinib (HS-10296) resensitizes ABCB1-overexpressing multidrug-resistant cancer cells to chemotherapeutic drugs Biochem Pharmacol 188 114516 https://doi.org/10.1016/j.bcp.2021.114516 PMID: 33713643 PMCID: 8291035
77. Fenn K, Maurer M, and Lee SM, et al (2020) Phase 1 study of erlotinib and metformin in metastatic triple-negative breast cancer Clin Breast Cancer 20(1) 80–86 https://doi.org/10.1016/j.clbc.2019.08.004 PMCID: 7304226
78. Li P, Zhang Q, and Torossian A, et al (2012) Simultaneous inhibition of EGFR and PI3K enhances radiosensitivity in human breast cancer Int J Radiat Oncol Biol Phys 83(3) e391–e397 https://doi.org/10.1016/j.ijrobp.2011.12.090 PMID: 22414288
79. El Guerrab A, Bamdad M, and Bignon YJ, et al (2020) Co-targeting EGFR and mTOR with gefitinib and everolimus in triple-negative breast cancer cells Sci Rep 10(1) 6367 https://doi.org/10.1038/s41598-020-63310-2 PMID: 32286420 PMCID: 7156377
80. Polychronis A, Sinnett HD, and Hadjiminas D, et al (2005) Preoperative gefitinib versus gefitinib and anastrozole in postmenopausal patients with oestrogen-receptor positive and epidermal-growth-factor-receptor-positive primary breast cancer: a double-blind placebo-controlled phase II randomised trial Lancet Oncol 6(6) 383–391 https://doi.org/10.1016/S1470-2045(05)70176-5 PMID: 15925816
81. Savage P, Blanchet-Cohen A, and Revil T, et al (2017) A targetable EGFR-dependent tumor-initiating program in breast cancer Cell Rep 21(5) 1140–1149 https://doi.org/10.1016/j.celrep.2017.10.015 PMID: 29091754
82. Zampino MG, Magni E, and Santoro L, et al (2008) Epidermal growth factor receptor serum (sEGFR) level may predict response in patients with EGFR-positive advanced colorectal cancer treated with gefitinib? Cancer Chemother Pharmacol 63(1) 139–148 https://doi.org/10.1007/s00280-008-0722-x PMID: 18327586
83. Costa R, Shah AN, and Santa-Maria CA, et al (2017) Targeting epidermal growth factor receptor in triple negative breast cancer: new discoveries and practical insights for drug development Cancer Treat Rev 53 111–119 https://doi.org/10.1016/j.ctrv.2016.12.010 PMID: 28104566
84. Sabatier R, Lopez M, Guille A, et al (2019) High response to cetuximab in a patient with EGFR-amplified heavily pretreated metastatic triple-negative breast cancer JCO Precis Oncol (3) 1–8
85. Diluvio G, Del Gaudio F, Giuli MV, et al (2018) NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest Oncogenesis 7(5) 42 https://doi.org/10.1038/s41389-018-0051-9 PMID: 29795369 PMCID: 5968025
86. Mehrabi M, Mansouri K, and Soleymani B, et al (2017) Development of a human epidermal growth factor derivative with EGFR-blocking and depleted biological activities: a comparative in vitro study using EGFR-positive breast cancer cells Int J Biol Macromol 103 275–285 https://doi.org/10.1016/j.ijbiomac.2017.05.035 PMID: 28501600
87. Zhang C, Yuan W, Wu Y, et al (2021) Co-delivery of EGFR and BRD4 siRNA by cell-penetrating peptides-modified redox-responsive complex in triple negative breast cancer cells Life Sci 266 118886 https://doi.org/10.1016/j.lfs.2020.118886