Lung cancer: a new frontier for microbiome research and clinical translation
Luis AJ Mur1, Sharon A Huws2, Simon JS Cameron3, Paul D Lewis4 and Keir E Lewis5,6
1Institute of Biological, Environmental and Rural Sciences, Aberystwyth University, Penglais Campus, Aberystwyth SY23 2DA, UK
2Institute for Global Food Security, School of Biological Sciences, Medical Biology Centre, Queen’s University Belfast, 97 Lisburn Road, Belfast BT9 7BL, UK
3Division of Computational and Systems Medicine, Department of Surgery and Cancer, Imperial College London, Charing Cross Hospital Campus, London W6 8RD, UK
4College of Medicine, Swansea University, Swansea SA2 8PP, UK
5Respiratory Unit, Prince Philip Hospital, Llanelli SA14 8QF, UK
6School of Medicine, University of Wales Swansea, Swansea SA2 8PP, UK
Correspondence to: Luis AJ Mur. Email: lum@aber.ac.uk
Abstract
The lung microbiome has been shown to reflect a range of pulmonary diseases—for example: asthma, chronic obstructive pulmonary disease (COPD) and cystic fibrosis. Studies have now begun to show microbiological changes in the lung that correlate with lung cancer (LC) which could provide new insights into lung carcinogenesis and new biomarkers for disease screening. Clinical studies have suggested that infections with tuberculosis or pneumonia increased the risk of LC possibly through inflammatory or immunological changes. These have now been superseded by genomic-based microbiome sequencing studies based on bronchoalveolar lavage, sputum or saliva samples. Although some discrepancies exist, many have suggested changes in particular bacterial genera in LC samples particularly, Granulicatella, Streptococcus and Veillonella. Granulicatella is of particular interest, as it appeared to show LC stage-specific increases in abundance. We propose that these microbial community changes are likely to reflect biochemical changes in the LC lung, linked to an increase in anaerobic environmental niches and altered pyridoxal/polyamine/nitrogenous metabolism to which Granulicatella could be particularly responsive. These are clearly preliminary observations and many more expansive studies are required to develop our understanding of the LC microbiome.
Keywords: lung cancer, microbiome, Granulicatella, ATP
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Published: 05/09/2018; Received: 25/04/2018
The lung microbiome
The concept that humans have ‘another genome’ in the associated microbiome is now firmly established [1]. Although not ignored, the lung microbiome has received less attention compared with the gut which could be a relic of the historical view that the normal lung is free from bacteria [2]. In fact, with typical air exchanges as expressed in the respiratory minute volume of 6 L (0.5 L × 12 breaths/minute), the upper and lower respiratory tracts are major sites of microbial exchanges with the environment [3]. Indeed, it is believed that between 1,500 and 14,000 microbes are inhaled each hour and the composition of the healthy lung microbiome is likely to reflect relative immigration, elimination through mechanisms such as coughing and microbial growth [4]. Within the healthy lung, epithelial layers are covered with only a thin mucosal layer with only <100 mL of mucus being produced per day; but are rich in lipid-rich surfactant to prevent alveolar collapse [5]. Therefore, the lung is a relatively low-nutrient environment for microbes. The lung microbiome is also distinctive to that of the upper respiratory tract [6] which displays greater similarities to that found in the stomach [7], although there are distinctive oral microbiomes reflecting discrete habitats [8].
The work of Hilty et al [6] represents a foundational study in lung microbiomics. Based on bronchial lavages of 43 patients, containing healthy individuals and those suffering from either chronic obstructive pulmonary disease (COPD) or asthma they found that seven genera dominated the lung, namely, Corynebacterium, Prevotella, Staphylococcus, Streptococcus, Veilonella, Haemophilus and Neisseria as defined by 16S rRNA gene sequencing. The lung microbiome is now a well-established focus of research into asthma, COPD and cystic fibrosis (CF). Nonetheless, there remain some technical questions regarding how to effectively sample the lung microbiome. Many studies have concentrated on bronchial lavages that are saline washes of parts of the lung obtained using a bronchoscope (e.g., [6, 7, 9]). This minimises the risk of contamination from the upper respiratory tract but clearly causes considerable patient discomfort. Sputum represents an alternative sampling route which, although representing an increased risk of contamination, can provide important insights into the lung microbiome [4].
Most likely due to its poor nutrient status, the number of bacteria is relatively low in healthy lungs but can increase markedly with respiratory disease [4]. This can reflect changes in pH, oxygen tension, temperature but perhaps most importantly changes in the adaptive and innate defence responses often linked to inflammatory events [10]. These are often linked to a substantial increase in the production of mucus which can represent an increased microbial nutrient source and this can be supplemented by free adenosine triphosphate [11] to encourage the growth of certain microbial species [4]. Considerations of which microbial changes occur during a range of lung pathologies have been the focus of many recent studies, which feature progressive worsening of chronic disease with occasional marked symptom exacerbations.
In asthma, transient incursions from Streptococcus are good predictors of asthma [12]. With CF disproportionately affects lung function due to a substantial build-up mucus, inflammation and microbial changes particularly Staphylococcus aureus, Haemophilus influenzae and Pseudomonas aeruginosa [13]. Microbiomic studies have suggested that some fermentative anaerobes are important to CF exacerbations [14]. COPD is a progressive disease associated with increasing shortness of breath and sputum production. Although the microbiome does not appear to change during the early stages of COPD as the disease advances there appear to be shifts towards genera which include pathogenic strains of Streptococcus, Pseudomonas and Haemophilus [15, 16]. The presence of pathogenic bacterial species is likely to be most significant during COPD exacerbations [17].
Microbial associations with lung cancer
Lung cancer (LC) remains a major global challenge, still responsible for 1.3 million deaths each year due to poor survival rates post-diagnosis. These relatively poor survival rates, compared to other cancers, are primarily a result of the late detection of a malignancy, where the success rates of clinical interventions are significantly reduced. LC exists in many histological types, each linked to various aetiology, developmental patterns and prognoses [18]. Whilst a role for the microbiome in LC does not seem to be immediately obvious compared to diseases such as CF or COPD, inflammatory responses are also a feature of LC [19] and this must be a starting point for considering a role for the microbiome.
Smoking is one of the prime risk factors in cancer and the associated carcinogens can readily form DNA adducts to promote genomic mutation. Equally, smoking can lead to chronic inflammation [20] and this may be the source of possibly 25% of all cancers [21, 22]. Mechanistically, this may arise from leukocytes producing reactive oxygen species, nitric oxide, metalloproteinases, interleukins and interferons, which could contribute to carcinogenesis by promoting increased genomic instability [23] Furthermore, cyclooxygenase (COX)-derived prostaglandin E2 can induce both tumour growth and metastasis. Given this, it has been shown that COX-inhibiting aspirin could suppress the development of LC although considerable heterogeneity amongst different sub-populations was observed [24].
If LC can arise from proinflammatory events, could chronic microbial infections could contribute to such as LC or respond to associated changes in the malignant lung? Such a link is apparently strong for a range of other cancers, especially for Helicobacter pylori and gastric cancer [25]. Here, the bacterial production of the CagA and some other 27–31 genes in a 40 kb gene cluster known as a pathogenicity island elicits a pronounced inflammatory response [26]. Other examples of bacterial infections linked to cancer are known [27] and are also referred to in other reviews in this edition of eCancer. In addition, the literature continues to provide some good indications that chronic lung diseases, such as tuberculosis (TB), as well as acute infection with pneumonia could be linked to lung carcinogenesis.
TB is the world’s leading cause of death from an infectious disease and been linked to around 1.4 million deaths in 2015 [28]. The causal agent is Mycobacterium tuberculosis (M. tuberculosis), which the following inhalation will infect and replicate within endosomes occurring in alveolar macrophages. Attempts by the macrophage to neutralise the bacterium are thwarted by the mycolic acid-based bacterial capsule. Once established, infected macrophages, T lymphocytes, B-lymphocytes and fibroblasts form a granuloma. More macrophages are recruited and fused to form multinucleated giant cells or lipid-rich foamy cells in the maturing granuloma. Crucially, many of these features are pro-inflammatory, and indeed, Tumour Necrosis Factor (TNF-a) and interferon (IFN-g) play an important role in driving granuloma formation. The granuloma, however, could be considered a failed form of host defence as M. tuberculosis may lie in a latent form for many years. The granuloma will eventually disintegrate allowing the bacteria to spread and form new lesions. However, about 90% of people infected with M. tuberculosis have asymptomatic, latent M. tuberculosis infections which very rarely activate to form an active disease [29].
Given the potential for TB infections to be foci for inflammation a possible link with LC was suspected but only recently has been supported by clinical studies [30]. A recent Danish study compared the cancer risk in TB patients during 1978–2011 with the general population and noted a significantly increased risk of LC [48]. Such an increased risk of LC in TB patients was also suggested in studies from Taiwan [31] and South Korea [32]. Histological studies have identified when LC and TB were coincident or where tumours formed from areas of pulmonary fibrosis linked to older sites of TB infection (TB scars) or from tuberculous cavities [33]. Extensive fibrosis may be associated with recurring TB infections and could decrease the clearance of lung lymph and lymph-associated particles. These have been linked to suppression of p21 to promote exit from Go in the cell cycle, increased expression of Bcl-2 will reduce apoptosis leading to genomic instability and finally, pro-inflammatory activation of cyclooxygenase 2 (COX-2) to produce eicosanoids which encourage angiogenesis [34].
Such observations are highly suggestive but a causal link is still uncertain as the immunosuppressive status seen with LC and its treatment could reactivate TB [35] so that this could bias any observations. Also, the possible role of latent TB infections, by far the most common form and carcinogenesis in the lung is unknown. Furthermore, it is unknown if inflammatory events linked to other chronic infection events could also be associated with LC. Thus, the SYNERGY project collected data from Canadian and EU patients with a chronic bronchitis, emphysema, TB and pneumonia which were tested for association with LC [36]. Interestingly, this study found no relationship between TB and LC. In contrast, a positive relationship was observed between LC and pneumonia, if it was diagnosed 2 years previously. However, no such association was seen in the female group which could reflect lower degrees of smoking. The strongest relationship was observed in patients where chronic bronchitis was present with either emphysema or pneumonia. In another population-based study in Taiwan, the incidence rate of LC was significantly higher in pneumococcal pneumonia patients than in controls with a striking hazard ratio of 4.24 [37].
The lung cancer microbiome
Given the observations of possible associations with microbial, pulmonary disease and LC, it is clear that microbiome investigations could provide some novel insights and potential biomarkers [38]. An initial retrospective study of bronchial aspirations collected by bronchoscopic endoscopy (n = 216), reported increases in potential pathogens of Gram-negative bacilli; including H. influenzae, Enterobacter sp., Escherichia coli as well as the Gram-positive Mycobacteria [39]. Metataxonomic 16S rRNA gene sequencing approaches were used by Lee et al [40] and 28 patients (20 LC, 8 benign biomass controls), again using bronchoalveolar lavage fluid (BALF). Two genera (Veillonella and Megasphaera) were significantly increased in LC patients and together could predict LC with an accuracy of 0.888 (as indicated with an area under the curve) [40]. In the following year, a paper was published on the salivary microbiome in patients (n = 10 in each category) with squamous cell carcinoma, adenocarcinoma and controls using targeted QPCR and 16S rRNA gene sequencing-based metataxonomics. They demonstrated that the salivary microbiome was different in the saliva of LC patients, particularly with respect to Capnocytophaga, Selenemonas, Veillonella and Neisseria. Using targeted QPCR they confirmed that Capnocytophaga and Veillonella were more abundant in LC patients, whilst Neisseria was conversely less abundant, suggesting their potential use as a bacterial biomarker. Of particular interest is the fact that in combination these could distinguish squamous cell carcinoma or adenocarcinoma from controls with significant but varying accuracies [41].
In another publication, the LC microbiome was investigated using paired samples of bronchial brushings from 24 LC patients one from the cancerous site, paired with a nearby noncancerous site and 18 from healthy controls from bronchoscopies [42]. They showed that the LC site exhibited decreased microbial diversity. However, the LC sites were enriched in Streptococcus compared to the controls which could be used to predict LC. Interestingly, this pair-sampling approach in LC patients suggested that the cancerous and non-cancerous microbiome were not very dissimilar which would argue for the LC microbiome reflecting wide-ranging changes in the lung [42].
Household pollution linked to burning coal may also be an LC risk factor which could be linked to the production of polycyclic aromatic hydrocarbons. A microbiomic component was tested in a never-smoked population of women from China with LC. This small-scale study revealed significant increases in Granulicatella, Abiotrophia and Streptococcus within sputum compared with the sputum of healthy controls [43]. Our own studies into the lung microbiome applied a full metagenomic approach to allow more insights into the LC microbiome in sputum [44]. Although this was a pilot study (n = 10), we noted increases in the range of species and in particular, Granulicatella adiacens which aligned with the study of Hosgood et al [43] . The increases in G. adiacens appeared to be particularly important as these correlated with the LC stage. As we have recently also published a metagenomic study of the sputum of COPD patients [16], we could compare the two pathologies compared to controls (Figure 1). To ease comparison with 16S rRNA amplicon-based studies, the heat map in Figure 1 focuses only significant changes at the level of genus. This indicated the increases in Granulicatella to be specific to LC (P = < 0.001, FDR 0.0004), but there were also significant increases in Mycobacterium (P = 0.0044, FDR 0.0133).
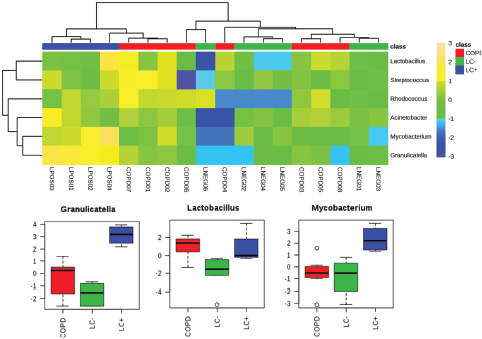
Figure 1. Significantly significant features of the LC microbiome which are observed in the sputum from patients with LC or COPD. We have previously published metagenomics studies of LC and COPD patients [44, 48]. Working at the level of ‘genus’, those showing significant differences between the microbiomes from LC (LC ) patients to patients referred to respiratory clinics, which were not found to have LC (LC−) or to COPD patients (further experimental details are found in [44, 48]), were extracted and compared using a heat map. Three genera showed significant increases in the LC category and these are presented in box and whisker format to depict log10 microbial abundance data.
Discussion: What do these initial studies tell us?
These are clearly early days in the field of LC microbiomics but there appears to be some commonality in the bacterial genera which are changing in LC patients with different authors reporting changes in Granulicatella [43, 44], Streptococcus [42–44] and Veillonella [40, 41]. Granulicatella species are a normal component of the upper respiratory tract [45] but can also cause some serious infections such as endocarditis [46]. Granulicatella is part of the Lactobacilliales order within the Streptococcaceae (including the genus Streptococcus), which also includes the family Enterococcaceae where the Enterococcus genus is located [47]. Within this genus, we also observed the change in sputum from patients with LC compared to patients referred to a respiratory clinic but proved not to have cancer−. Therefore, it could be suggested that there seems to be some similarity in microbial responses to LC. Streptococcus, Enterococcus and Granulicatella are facultative anaerobes and the detection of Veillonella as an obligate anaerobe would argue that LC is associated with a switch to anaerobic fermentative respiration. In this context, it is relevant that the fermentative facultative anaerobe Lactobacillus is significantly increased in both COPD and LC patients where biofilm formation is prominent. However, the specificity that we have seen with Granulicatella would argue a specific microbiomic response to LC. Granulicatella is considered to be an example of nutritionally variant Streptococci as it requires such as pyridoxal (a form of vitamin B6) or L-cysteine to be supplemented to media in order to grow in the laboratory [46]. Our metagenomics strategy suggested functional changes in the LC microbiome which could be relevant to the increases in Granulicatella. In particular, it indicated increases in the microbiome's capacity to metabolise putrescine and its polyamine products [44]. This tallied with the increases in putrescine in LC patients detected in our parallel metabolomic study [48]. As polyamine and pyridoxal metabolism can be intrinsically linked, it may be that the increased abundance of Granulicatella could be a response to metabolites in these pathways [49]. Deregulation of putrescine and polyamines, which influence the cell cycle, has been noted in several cancers; including lung [50]. Therefore, Granulicatella could be responding to LC-linked increases in these metabolites.
Such considerations suggest that the microbiome reflects changes in the cancerous lung environment, but a causative role cannot be ruled out. The various mechanisms through which the microbiome could be causative in LC have been reviewed by Mao et al [38] and include inflammatory and immune changes which could reflect microbiological dysbiosis so that pathogens begin to predominate. In this context, the increases in genera with important pathogens, Streptococcus and Mycobacterium (Figure 1), are relevant but these changes have not been observed in all studies. Here, comparisons with COPD may be informative where associations with a high prevalence of P. aeruginosa, H. influenzae and S. pneumoniae indicate an unequivocal role for pathogens [4, 16].
Taking all of our points together, there is a clear requirement for a larger scale assessment of microbiome changes in LC. Such studies should be sensitive to the heterogeneity of LC in terms of histology and stage and either focus on particular examples or be large enough to have the statistical power to allow valid conclusions to be made. A single sampling technique should be used, which although may be better done using BALF, restricts the number of samples which can be taken due to the invasive nature of this technique, and as such sputum samples may be a better alternative allowing larger numbers of samples to be obtained. Sputum also appears to retain the features of the lower respiratory tract. Finally, a full metagenomic strategy rather than 16S rRNA gene amplicon sequencing would allow functional changes in the microbiome to be revealed. Ideally, these could be supplemented with metatranscriptomic, metaproteomic or metabolomic assessments of samples so that these functional changes can be related to the wider lung environment. The challenge is considerable, but it does offer the possibility of yielding new insights into lung carcinogenesis, new biomarkers and possibly chemotherapeutic drug metabolism.
Conclusions
The lung microbiome is now a well-established feature of many types of respiratory diseases. Preliminary microbiome analyses have indicated some changes in genera which include pathogens that are significantly associated with LC. These could indicate a role for microbially associated pro-inflammatory or immunological events in LC. Equally, the microbial changes could reflect changes in the LC environment to which the microbiome is responding. Whatever the cause, specific microbial changes could represent specific biomarkers for LC presence and type. However, the paucity of the number of LC microbiomic studies, their sample sizes and the range of sampling techniques used prevent any firm conclusions being made. More and larger studies are required.
Acknowledgments
The authors acknowledge the support of an Aberystwyth University Postgraduate scholarship to SJSC. The data presented here was part of The MedLung observational study (UKCRN ID 4682) and is covered by loco-regional ethical approval from the Hywel Dda Health Board, UK (05/WMW01/75). This work was made possible through BBSRC funded infrastructure at the IBERS, UK.
Authors’ contributions
All the authors wrote and edited the review. The representation of previously published data was based on the work of SJSC. The Medlung study was led by PDL and KL.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
1. Zhao LP (2010) GENOMICS The tale of our other genome Nature 465(7300) 879–880 https://doi.org/10.1038/465879a PMID: 20559375
2. Cotran RS, Kumar V, and Collins T, et al (1999) Robbins Pathologic Basis of Disease (Philadelphia: Saunders) pp 87–118
3. McArdle WD, Katch FI, and Katch VL (2006) Essentials of Exercise Physiology (Baltimore, MD: Lippincott Williams & Wilkins) pp 265–280
4. Dickson RP, Erb-Downward JR, and Martinez FJ, et al (2016) The microbiome and the respiratory tract Annu Rev Physiol 78 481–504 https://doi.org/10.1146/annurev-physiol-021115-105238 PMCID: 4751994
5. Wu HX, Kuzmenko A, and Wan SJ, et al (2003) Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability J Clin Invest 111(10) 1589–1602 https://doi.org/10.1172/JCI16889 PMID: 12750409 PMCID: 155045
6. Hilty M, Burke C, and Pedro H, et al (2010) Disordered microbial communities in asthmatic airways PLoS One 5(1) e8578 https://doi.org/10.1371/journal.pone.0008578 PMID: 20052417 PMCID: 2798952
7. Bassis CM, Erb-Downward JR, and Dickson RP, et al (2015) Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals Mbio 6(2) e00037 https://doi.org/10.1128/mBio.00037-15 PMID: 25736890 PMCID: 4358017
8. Dewhirst FE, Chen T, and Izard J, et al (2010) The human oral microbiome J Bacteriol 192(19) 5002–5017 https://doi.org/10.1128/JB.00542-10 PMID: 20656903 PMCID: 2944498
9. Dickson RP, Martinez FJ, and Huffnagle GB (2014) The role of the microbiome in exacerbations of chronic lung diseases Lancet 384(9944) 691–702 https://doi.org/10.1016/S0140-6736(14)61136-3 PMID: 25152271 PMCID: 4166502
10. Dickson RP, Erb-Downward JR, and Huffnagle GB (2014) Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis Lancet Respir Med 2(3) 238–246
11. Schmidt A, Belaaouaj A, and Bissinger R, et al (2014) Neutrophil elastase-mediated increase in airway temperature during inflammation J Cyst Fibros 13(6) 623–631 https://doi.org/10.1016/j.jcf.2014.03.004 PMID: 24713593
12. Teo SM, Mok D, and Pham K, et al (2015) The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development Cell Host Microbe 17(5) 704–715 https://doi.org/10.1016/j.chom.2015.03.008 PMID: 25865368 PMCID: 4433433
13. Mogayzel PJ, Naureckas ET, and Robinson KA, et al (2013) Cystic fibrosis pulmonary guidelines chronic medications for maintenance of lung health Am J Resp Crit Care 187(7) 680–689 https://doi.org/10.1164/rccm.201207-1160OE
14. Magalhaes AP, Azevedo NF, and Pereira MO, et al (2016) The cystic fibrosis microbiome in an ecological perspective and its impact in antibiotic therapy Appl Microbiol Biot 100(3) 1163–1181 https://doi.org/10.1007/s00253-015-7177-x
15. Erb-Downward JR, Thompson DL, and Han MK, et al (2011) Analysis of the Lung Microbiome in the ‘Healthy’ Smoker and in COPD PLoS One 6(2) e16384 https://doi.org/10.1371/journal.pone.0016384 PMID: 21364979 PMCID: 3043049
16. Cameron SJS, Lewis KE, and Huws SA, et al (2016) Metagenomic sequencing of the chronic obstructive pulmonary disease upper bronchial tract microbiome reveals functional changes associated with disease severity PLoS One 11(2) e0149095 https://doi.org/10.1371/journal.pone.0149095 PMID: 26872143 PMCID: 4752236
17. Huang YJ, Kim E, and Cox MJ, et al (2010) A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations Omics 14(1) 9–59 https://doi.org/10.1089/omi.2009.0100 PMID: 20141328 PMCID: 3116451
18. (UK) NCCfC (2011) The Diagnosis and Treatment of Lung Cancer (Update) (Cardiff: National Collaborating Centre for Cancer (UK))
19. Cho WC, Kwan CK, and Yau S, et al (2011) The role of inflammation in the pathogenesis of lung cancer Expert Opin Ther Targets 15(9) 1127–1137 https://doi.org/10.1517/14728222.2011.599801 PMID: 21751938
20. Walser T, Cui X, and Yanagawa J, et al (2008) Smoking and lung cancer: the role of inflammation Proc Am Thorac Soc 5(8) 811–815 https://doi.org/10.1513/pats.200809-100TH PMID: 19017734 PMCID: 4080902
21. Balkwill FR and Mantovani A (2012) Cancer-related inflammation: common themes and therapeutic opportunities Semin Cancer Biol 22(1) 33–40 https://doi.org/10.1016/j.semcancer.2011.12.005 PMID: 22210179
22. Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation Cell 144(5) 646–674 https://doi.org/10.1016/j.cell.2011.02.013 PMID: 21376230
23. Chumduri C, Gurumurthy RK, and Zietlow R, et al (2016) Subversion of host genome integrity by bacterial pathogens Nat Rev Mol Cell Bio 17(10) 659–673 https://doi.org/10.1038/nrm.2016.100
24. Hochmuth F, Jochem M, and Schlattmann P (2016) Meta-analysis of aspirin use and risk of lung cancer shows notable results Eur J Cancer Prev 25(4) 259–268 https://doi.org/10.1097/CEJ.0000000000000176
25. Crowe SE (2005) Helicobacter infection, chronic inflammation, and the development of malignancy Curr Opin Gastroenterol 21(1) 32–38 PMID: 15687882
26. Ahn HJ and Lee DS (2015) Helicobacter pylori in gastric carcinogenesis World J Gastrointest Oncol 7(12) 455–465 https://doi.org/10.4251/wjgo.v7.i12.455 PMID: 26690981 PMCID: 4678392
27. Khan AA and Shrivastava A (2010) Bacterial infections associated with cancer: possible implication in etiology with special reference to lateral gene transfer Cancer Metast Rev 29(2) 331–337 https://doi.org/10.1007/s10555-010-9217-4
28. WHO (2017) Global Tuberculosis Report 2017 (Geneva: WHO Press)
29. Fogel N (2015) Tuberculosis: a disease without boundaries Tuberculosis 95(5) 527–531
30. Mejri I, Ourari B, and Cherif H, et al (2016) Pulmonary tuberculosis and lung cancer: a complex interaction Eur Respir J 48 (suppl 60)
31. Su VY, Yen YF, and Pan SW, et al (2016) Latent tuberculosis infection and the risk of subsequent cancer Medicine (Baltimore) 95(4) e2352 https://doi.org/10.1097/MD.0000000000002352
32. Bae JM, Li ZM, and Shin MH, et al (2013) Pulmonary tuberculosis and lung cancer risk in current smokers: the Seoul Male Cancer Cohort Study J Korean Med Sci 28(6) 896–900 https://doi.org/10.3346/jkms.2013.28.6.896 PMID: 23772155 PMCID: 3678007
33. Cukic V (22017) The association between lung carcinoma and tuberculosis Med Arch 71(3) 212–214 PMID: 28974836 PMCID: 5585804
34. Ardies CM (2003) Inflammation as cause for scar cancers of the lung Integr Cancer Ther 2(3) 238–246 https://doi.org/10.1177/1534735403256332
35. Kamboj M and Sepkowitz KA (2006) The risk of tuberculosis in patients with cancer Clin Infect Dis 42(11) 1592–1595 https://doi.org/10.1086/503917 PMID: 16652317
36. Denholm R, Schuz J, and Straif K, et al (2014) Is previous respiratory disease a risk factor for lung cancer? Am J Resp Crit Care 190(5) 549–559 https://doi.org/10.1164/rccm.201402-0338OC
37. Lin TY, Huang WY, and Lin JC, et al (2014) Increased lung cancer risk among patients with pneumococcal pneumonia: a Nationwide Population-Based Cohort Study Lung 192(1) 159–165 https://doi.org/10.1007/s00408-013-9523-z
38. Mao QX, Jiang F, and Yin R, et al (2018) Interplay between the lung microbiome and lung cancer Cancer Lett 415 40–48 https://doi.org/10.1016/j.canlet.2017.11.036
39. Laroumagne S, Salinas-Pineda A, and Hermant C, et al (2011) Incidence and characteristics of bronchial colonisation in patient with lung cancer: a retrospective study of 388 cases Rev Mal Respir 28(3) 328–335
40. Lee SH, Sung JY, and Yong D, et al (2016) Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions Lung Cancer 102 89–95 https://doi.org/10.1016/j.lungcan.2016.10.016 PMID: 27987594
41. Yan XM, Yang MX, and Liu J, et al (2015) Discovery and validation of potential bacterial biomarkers for lung cancer Am J Cancer Res 5(10) 3111–3122 PMID: 26693063 PMCID: 4656734
42. Liu HX, Tao LL, and Zhang J, et al (2018) Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects Int J Cancer 142(4) 769–778 https://doi.org/10.1002/ijc.31098
43. Hosgood HD, Sapkota AR, and Rothman N, et al (2014) The potential role of lung microbiota in lung cancer attributed to household coal burning exposures Environ Mol Mutagen 55(8) 643–651 https://doi.org/10.1002/em.21878 PMID: 24895247 PMCID: 4217127
44. Cameron SJS, Lewis KE, and Huws SA, et al (2017) A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer PLoS One 12(5) e0177062
45. Aas JA, Paster BJ, and Stokes LN, et al (2005) Defining the normal bacterial flora of the oral cavity J Clin Microbiol 43(11) 5721–5732 https://doi.org/10.1128/JCM.43.11.5721-5732.2005 PMID: 16272510 PMCID: 1287824
46. Cargill JS, Scott KS, and Gascoyne-Binzi D, et al (2012) Granulicatella infection: diagnosis and management J Med Microbiol 61(6) 755–761 https://doi.org/10.1099/jmm.0.039693-0 PMID: 22442291
47. Ludwig W, Schleifer K, and Whitman WB (2009) Family IV. Enterococcaceae fam nov 2nd edn, eds Vos P, Garrity G, and Jones D, et al (New York: Springer)
48. Cameron SJ, Lewis KE, and Beckmann M, et al (2016) The metabolomic detection of lung cancer biomarkers in sputum Lung Cancer 94 88–95 https://doi.org/10.1016/j.lungcan.2016.02.006 PMID: 26973212
49. Pegg AE (1977) Role of pyridoxal-phosphate in mammalian polyamine biosynthesis—lack of requirement for mammalian s-adenosylmethionine decarboxylase activity Biochem J 166(1) 81–88
50. Thomas T and Thomas TJ (2003) Polyamine metabolism and cancer J Cell Mol Med 7(2) 113–126 https://doi.org/10.1111/j.1582-4934.2003.tb00210.x PMID: 12927050


