Current advances in cancer immunohistochemistry: a new perspective for the Ki-67 biomarker
Talita Alves do Nascimento Santos1, Anna Karoline Fausto da Silva2, Karin Soares Gonçalves Cunha3, Camila Braz Pereira da Costa4,5, Aldo Rodrigues da Silva1,5, Helena Carla Castro1,5 and Nathália Silva Carlos Oliveira3
1Programa de Pós-Graduação em Patologia, Faculdade de Medicina, Hospital Universitário Antônio Pedro, Universidade Federal Fluminense, Niteroi, RJ 24033-900, Brazil
2Serviço de Anatomia Patológica, Hospital Universitário Antônio Pedro, Universidade Federal Fluminense, Niteroi, RJ 24033-900, Brazil.
3Departamento de Patologia, Faculdade de Medicina, Hospital Universitário Antônio Pedro, Universidade Federal Fluminense, Niteroi, RJ 24033-900, Brazil
4Diretoria Industrial, Instituto Vital Brazil, Niteroi, RJ 24230-410, Brazil
5LABiEMOL - PPBI - Departamento de Biologia Molecular e Celular, Instituto de Biologia, Universidade Federal Fluminense, Niteroi, RJ 24210-201, Brazil
Abstract
Ki-67 is a cell proliferation biomarker used to evaluate the proliferative activity of neoplasia cells. However, considering its functions on the cell cycle, the standard method seems to be an underused way of evaluating expression, since so far, its analytical validity of Ki-67 remains questionable for its use in personalised therapy. Improvements in the assessment of Ki-67 expression continue to be explored, and recently, a new approach that considers the heterogeneity or variability in staining intensity has emerged as a more improved way than the traditional method. In this review, we bring together what is available in the literature on the biological properties of the protein and highlight how this potential association is promising in the field of personalised medicine.
Keywords: Ki-67, immunohistochemistry, cell proliferation, biomarker, immunostaining, nuclear protein, personalised medicine, neoplasia, histopathological diagnostic
Correspondence to: Nathália Silva Carlos Oliveira
Email: ns_oliveira@id.uff.br
Published: 05/03/2025
Received: 16/09/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Trial registration
Since this is a narrative review with no meta-analysis, it does not involve the conduction of new clinical trials, patient recruitment or primary data collection. Therefore, it falls outside the scope of studies requiring trial registration in databases such ClinicalTrials.gov or WHOICTRP.
Introduction
Cell proliferation is a fundamental process critical for tissue homeostasis, driven by tightly regulated and coordinated events that ensure accurate segregation and transmission of genetic material to daughter cells. However, disruptions in the regulatory mechanisms of the cell cycle can lead to uncontrolled proliferation, resulting in pathological conditions such as neoplasms [1].
Malignant neoplasms are a significant global health challenge, with the World Health Organization reporting approximately 10 million deaths annually, making cancer the second leading cause of mortality worldwide [2]. Despite advancements in prevention, treatment strategies and understanding tumour biology, early diagnosis remains the most effective means to improve clinical outcomes and, unfortunately, many cases are diagnosed at advanced stages [3].
In this context, precision medicine [4] has emerged as a promising approach for personalizing therapies and improving patient survival [5]. Genes associated with cell proliferation are notably upregulated during the cell cycle, and the detection of their protein products has become an invaluable diagnostic and prognostic tool. Among these, the nuclear protein Ki-67 stands out. Expressed during all active phases of the cell cycle – except in resting cells and early G1 – it is widely used in anatomopathological analyses to assess the growth fraction of cell populations [6].
The utility of Ki-67 extends beyond its role in monitoring cell cycle activity. It is routinely detected through standard immunodetection techniques such as immunohistochemistry and immunocytochemistry [7]. The labelling index (LI), calculated as the percentage of Ki-67-positive cells relative to the total cell count in a sample, provides insights into tumour growth dynamics. Together with the mitotic index (the count of cells in mitosis), the LI offers estimates of tumour aggressiveness and malignancy, contributing to the classification of tumours [8]. Furthermore, Ki-67 expression can serve as a prognostic marker, indicating tumour progression or outcomes and a predictive marker, forecasting recurrence, progression or mortality risk [9].
The prognostic and predictive significance of Ki-67 has been established for several malignancies [10, 11], including non-Hodgkin lymphomas [12], renal carcinoma [13], soft tissue sarcomas [14], prostate carcinoma [15], breast cancer [9] and neuroendocrine tumours (NETs) [16]. Intriguingly, variations in both the intensity and nuclear distribution of Ki-67 expression have been observed during histopathological evaluations, though these heterogeneities are often overlooked in routine diagnostics [17].
Emerging evidence highlights that the functions of Ki-67 and nuclear localization vary depending on the cell cycle phase, offering a nuanced view of cell cycle dynamics [18]. This phase-specific distribution underscores its potential as a biomarker not just for proliferation but also for cell cycle status. Harnessing these characteristics could pave the way for personalised antitumour therapies, enabling phase-specific classifications of proliferative cells. This approach could refine the use of Ki-67 in modulating treatment strategies, enhancing its relevance in precision oncology.
KI-67: molecular and genetics characteristics
The Ki-67 protein is encoded by the MKI67 gene, located on chromosome 10q25.qter in humans. This gene spans approximately 29,965 base pairs and consists of 15 exons and 14 introns [19]. Its transcription produces multiple isoforms through alternative splicing, including Ki-67 α, β, δ, ε and γ [20]. Among these, the α and β isoforms are detected in cancer cells, with the β isoform being exclusively found in normal cells, linked to cell cycle progression [21]. The functions of other isoforms remain underexplored.
Structurally, Ki-67 includes distinct functional domains.
N-terminal domain: It features a forkhead-associated domain involved in chromatin regulation and early ribosomal RNA synthesis, along with a binding site for protein phosphatase 1, which modulates chromatin dynamics.
Central region: It contains 16 disordered repeats (‘Ki-repeats’) that may contribute to phase separation.
C-terminal domain: It contains a leucine-rich region essential for chromosomal association during mitosis [13].
The MKI67 promoter includes regulatory elements such as cell cycle-dependent elements (CDEs) and cell cycle homology regions (CHRs), typical of late cell cycle genes [22]. Although mRNA levels of MKI67 are similar across cell lines, transcription is phase-regulated [23]. The DREAM complex, which interacts with the CDE and CHR regions, plays a pivotal role in gene activation and repression. Specifically, the following conditions hold.
In G0 and early G1, the RBL1-DREAM-MuvB repressor complex binds the promoter and inhibits transcription [24].
After mitogenic signals, cyclin D-CDK4/6 phosphorylates RBL1, dissociating the repressor complex, but transcription remains inactive [24, 25].
At the G1/S transition, the MuvB complex recruits the BMyB protein, activating transcription [26].
In G2, further recruitment of FoxM1 to the MuvB-BMyB complex enhances transcription until mitosis [26].
The dynamics of MKI67 transcription differ between normal and cancerous cells [27]. For instance, MKI67 expression remains constant in cancer cells without evident mutations, but variations in transcription patterns during the S phase have been observed across different cell lines [27–31]. These discrepancies may arise from differences in experimental protocols or the pathways involved in G0-G1 and Mitosis-G1 transitions, warranting further investigation.
Protein kinetics also reveal that Ki-67 has a short half-life (~1 hour) [32], with degradation mediated by the anaphase-promoting complex (APC/C-Cdh1) in G0, early G1 and late mitosis [23]. Recent findings suggest that Ki-67 ubiquitination in post-mitotic neurons involves the APC7 subunit, potentially linking Ki-67 to neurological conditions [33]. Enhanced understanding of Ki-67 degradation and expression variability during mitosis and G1 could provide insights into its role in cell cycle regulation [33].
In conclusion, while the transcriptional regulation and structural domains of Ki-67 are increasingly understood, further studies are needed to elucidate its isoform-specific roles, variability in expression patterns and degradation mechanisms in normal and pathological conditions.
KI-67 function and distribution during the cell cycle
Ki-67 demonstrates dynamic localization and functional roles throughout the cell cycle, regulated by post-translational modifications, particularly phosphorylation. In the early interphase, Ki-67 is dephosphorylated, allowing DNA binding at satellite regions. As the nuclear bodies coalesce and nucleoli form, Ki-67 interacts with nucleolar protein NIFKh, relocating to the nucleolar cortex and forming a reticulated structure. During the S and G2 phases, Ki-67 mRNA transcription increases, with the protein accumulating in the nucleoli and nucleoplasm, particularly as cells approach mitosis [34].
At mitosis onset, cyclin B-Cdk1 phosphorylates Ki-67 at specific sites, altering its charge and dissociating it from DNA, facilitating its migration to the chromosome periphery to form the perichromosomal layer. This dynamic process, influenced by interactions with kinesin-like motor proteins, is reversed at mitosis completion, restoring nonphosphorylated state of Ki-67 and its DNA-binding capacity [34–37].
The localization of Ki-67 shifts during the cell cycle has been extensively documented using in vitro models [28, 30, 32, 34, 38–40]. During interphase, it organises heterochromatin [41], guides nucleolar organizing regions (NORs) [40] and participates in ribosome biogenesis [42, 43]. In mitosis, it aids in perichromosomal layer formation, chromosomal mobility and mitotic chromosome clustering [18, 35, 43, 44]. Morphological and functional variations are evident, such as Ki-67 granules in G1 and nucleoplasm-wide distribution in the late S phase [40, 41].
Notably, discrepancies in S and G2 phase patterns among cell lines suggest methodological influences or intrinsic variability in Ki-67 expression [17, 28]. Mitosis is the most consistently described phase, where Ki-67 localises to the chromosomal periphery [18, 43], facilitating chromosome segregation [39] and nucleolar reassembly [45]. The protein either returns to heterochromatin in cycling cells or degrades in quiescent cells [23]. Based on published data, distinct Ki-67 morphological patterns correlate with specific cell cycle phases, Figure 1, reinforcing its utility as a biomarker for cellular dynamics and therapeutic targeting.
KI-67: histopathological application and new analysis methods
The Ki-67 labelling index is a widely used method in histopathology to assess tumour proliferative activity and determine its aggressiveness. This method involves counting the number of Ki-67-positive cells relative to the total number of cells within a predefined area. The percentage of positive cells is calculated by multiplying the result by 100 and Ki-67 positive is often classified using cutoff points, which stratify proliferative activity as low, medium or high [46, 47]. This allows for the determination of malignancy status and, in some cases, provides prognostic or predictive value [48].
Although workgroups have developed guidelines for Ki-67 usage and specified cutoff points based on tumour type, these guidelines require regular review and updates [30, 47–49]. Traditionally, Ki-67 quantification relies on a binary system (positive or negative), but several alternative methods exist, including visual estimation using a microscope, manual counting of digital images and automated analysis using machine learning algorithms. However, a consensus on the most suitable method has yet to be established [49].
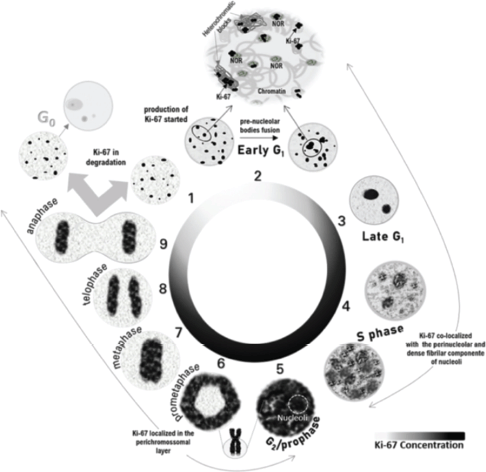
Figure 1. Schematic representation of Ki-67 localization throughout the cell cycle based on previously published experiments. During the early G1 phase, Ki-67 remains from the previous cell cycle located in the heterochromatic blocks and co-localises with the NORs. After the fusion of NORs, Ki-67 can be identified in the nucleoli. At the end of the G1 phase, the small nucleoli gather into one or two larger nucleoli and Ki-67 is localised in the nucleoli as discrete points in the nucleoplasm. At the beginning of the S phase, Ki-67 continues to be in the nucleoli and appears as coarser granules throughout the nucleoplasm, filling a large part of the nucleoplasm at the end of the S phase. During the G2 phase and until prophase, the nucleoli are barely evident due to the almost complete filling of the nucleoplasm by Ki-67, coinciding with the maximum expression of MKI67. From prometaphase to anaphase, Ki-67 is relocated to the structure of mitotic chromosomes. Cells that enter rest after mitosis have degraded Ki-67.
One challenge in validating Ki-67 analytically is the heterogeneity of nuclear immunostaining, which varies with its localization during the cell cycle. Traditional binary analyses overlook these variations, despite evidence from several studies showing that they provide valuable insights into the function of Ki-67 throughout the cell cycle [49, 50]. Recognizing this, recent research has proposed innovative approaches to evaluate Ki-67 expression, accounting for its graduated nature.
A pioneering study introduced a theoretical model linking nuclear patterns to cell cycle phases, classifying Ki-67 staining into five distinct patterns (NP1–NP5). This approach demonstrated that specific nuclear patterns correlate with cell cycle phases, providing a nuanced understanding of proliferative activity Table 1, Figure 2A. For example, NP1 was predominantly observed in tissues without lesions or with hyperplasia, indicative of cells in the G1 phase [17].
In contrast, carcinomas displayed higher proportions of cells with NP4 staining, representing the G2/M phase. The method showed high interobserver reliability and moderate diagnostic agreement. In addition, haematoxylin–eosin staining improved mitotic phase identification, except for prophase, which overlapped with G2 phase cells [17].
Another study focused on the intensity of Ki-67 staining in NET samples, disregarding localization. By considering only intensely stained nuclei (associated with the G2/M phase), this approach reduced false negatives seen in traditional methods. It resulted in up to a 30% reduction in high-grade tumour classifications and demonstrated potential predictive value for survival rates in NET patients [52] Table 1, Figure 2B.
A third study integrated nuclear morphometry, chromatin texture, staining intensity and localization to create a nuclear gradient classification system. This semiautomated method enabled the correlation of Ki-67 nuclear gradients with tumour activity and prognosis in pulmonary typical carcinoma Table 1, Figure 2C. High and low gradients were visualised using heat maps, underscoring the utility of this technique in diagnosis. While advanced techniques, such as flow cytometry, multiplex immunohistochemistry and FUCCI fluorescence, provide valuable cell cycle data, their limitations include high costs, time consumption and reliance on specialised equipment and expertise. Moreover, these techniques often fail to capture intratumoural heterogeneity due to extensive cellular manipulation [53].
Table 1. Comparison of classifications for immunohistochemistry cited by the authors.
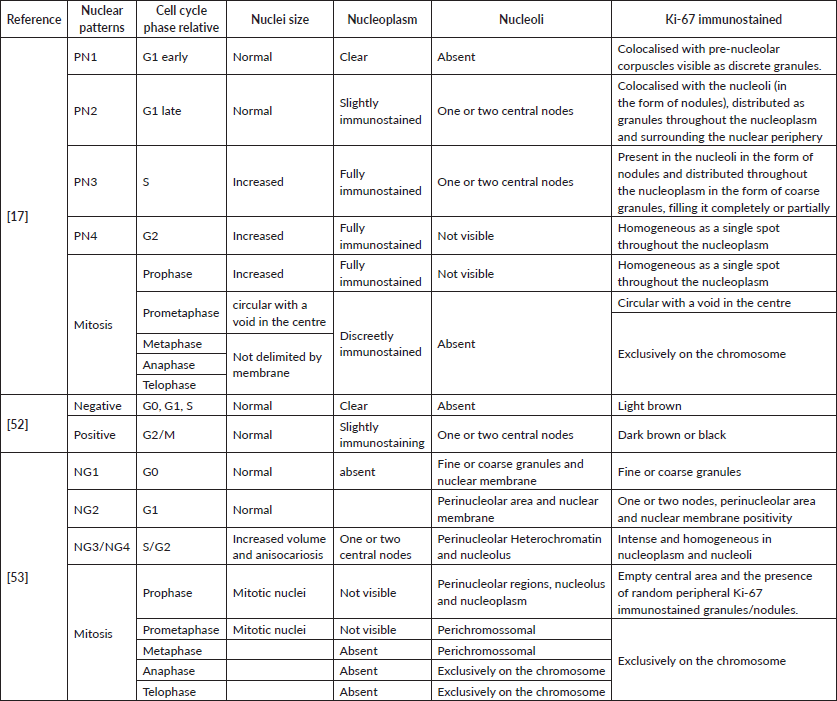
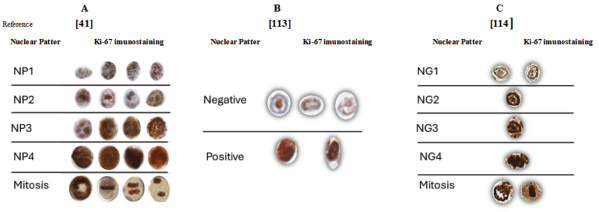
Figure 2. Comparison of the representation of Ki-67 patterns immunostaining. (a): Nuclear patterns categorised according to the cell cycle phases. (b): Nucleus categorised in negative or positive according to the immunostaining intensity. (c): Nucleus categorised by the nuclear gradient according to the cell cycle phases, chromatin texture and cell cycle phase.
An alternative method involves analysing DNA content with DAPI, which considers differences in DNA texture, content and shape to identify cell cycle phases. However, challenges remain in accurately distinguishing transitions between G1/S, S/G2 and G2/M phases. To address these limitations, further research into the relationship between Ki-67 morphological patterns and cell cycle phases in formalin-fixed paraffin-embedded samples holds promise. This approach leverages a widely used marker in routine histopathology, offering the potential for improved classification of cell cycle phases using a single, accessible marker.
Therefore, to enhance the practical applications of Ki-67 in histopathological analyses, beyond its conventional usage, new methodologies and approaches have emerged to refine its diagnostic and prognostic capabilities, and some of these uses and advantages are summarised in Table 2.
Table 2. List of uses and advantages of practical applications of Ki-67 in histopathological analyses.
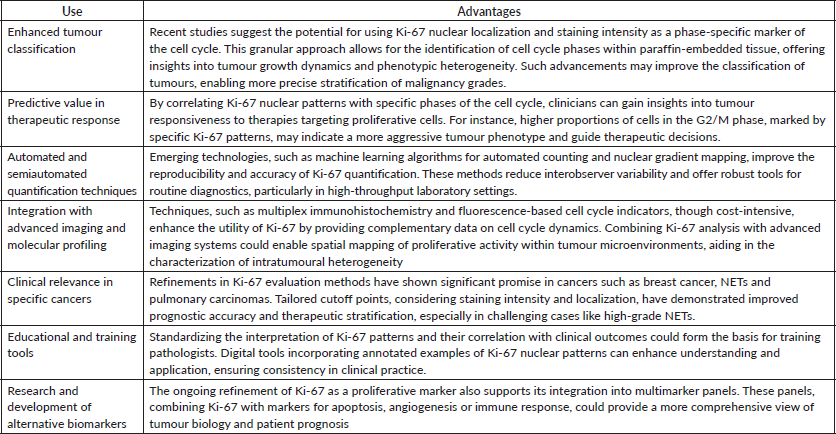
By integrating these advancements into routine practice, Ki-67 assessment not only remains a cornerstone of tumour biology but also evolves to meet the demands of precision medicine, ultimately improving diagnostic accuracy and patient outcomes. Therefore, we are faced with the possibility of classifying the phases of the cell cycle using a single marker, already known and widely used in routine histopathology (Table 2).
Discussion
In this review, we described studies that demonstrated new methods for semiquantitative evaluation of the immunohistochemical expression of Ki-67. It discusses innovative methods that account for the variability in Ki-67 expression and localization throughout the cell cycle, enabling the categorization of morphological patterns in formalin-fixed, paraffin-embedded tissues. The proposed classification systems incorporate morphological and morphometric characteristics, along with the heterogeneity of Ki-67 immunostaining. They can be broadly divided into two categories: one based on differences in protein localization and intensity during cell cycle phases and the other solely focused on variations in staining intensity [17, 52, 53].
The first approach highlights the biomarker potential for mapping cell cycle dynamics, while the second aims to mitigate the impact of immunostaining heterogeneity on tumour stratification. Both approaches demonstrated positive outcomes when compared to the gold standard, including improvements in histological classification, prognostic definition and clinical outcome prediction.
Mapping cell cycle dynamics is crucial, particularly in cancer – a complex and heterogeneous disease influenced by metabolic, genetic, epigenetic, molecular and microenvironmental factors. Tumour heterogeneity complicates classification, prognosis and therapeutic decisions, contributing to recurrence and metastasis [50]. As cancer is fundamentally a disorder of cell proliferation, cell cycle-specific chemotherapies represent a viable treatment strategy [51]. However, effective application requires accurate mapping of cell cycle dynamics [54, 55]. While immunohistochemistry is useful for cell cycle characterization, relying on specific markers for each phase is labour-intensive and costly [54]. Similarly, advanced techniques, such as fluorescence in situ hybridization, next-generation sequencing and single-cell sequencing, though valuable, are prohibitively expensive for routine diagnosis [56–58].
The proposed new methods simplify this process by leveraging a single, well-established biomarker, Ki-67, already widely used in routine histopathology. These methods offer advantages in time and cost, essential factors in reducing cancer-related morbidity and mortality. Improvements in Ki-67 evaluation have enhanced its diagnostic, prognostic and predictive value, presenting new opportunities for personalised therapy. Future studies should prioritise validating these methods in diverse tissue types, both normal and neoplastic, using multiplex immunohistochemistry to explore their full potential.
One challenge in Ki-67 quantification arises from overestimating positive nuclei when following WHO guidelines, which count any Ki-67-positive cell. This can lead to misclassification, as resting or slow-growing cells with residual Ki-67 may be erroneously included as actively proliferating [sobecki]. To address this, some studies propose a classification based on staining intensity, aiming to reduce false positives. While this approach has shown promise, especially in NET, it requires caution. Protein accumulation over multiple cell cycles poses challenges in setting accurate thresholds, yet this method may help identify slow-growing cells, which are associated with chemotherapy resistance, recurrence and metastasis.
Finally, understanding Ki-67 dynamics could bridge gaps in cancer characterization. By refining its application in personalised therapy, Ki-67 has the potential to become a robust clinicopathological biomarker. Future studies should focus on validating these innovative methods in formalin-fixed, paraffin-embedded tissues across various proliferative disorders. With its widespread use, low cost and ease of implementation, Ki-67 remains a promising tool for advancing histopathological and therapeutic precision.
Conclusion
In conclusion, this review highlights new methods proposed for analysing Ki-67 expression in routine histopathology. By considering the diverse nuclear patterns and distribution of Ki-67 across cell cycle phases, these methods demonstrated advantages over the traditional gold standard approach. Notably, they raise the possibility of mapping cell cycle dynamics by correlating Ki-67 patterns with specific phases. However, as these methods rely on empirical models, further research is required to explore their full potential.
Given that the theoretical foundation was derived from studies on cell lines and various techniques, it is recommended to validate these models in formalin-fixed, paraffin-embedded tissues. Applying multiplex immunohistochemistry to samples representing different proliferative states (normal,
hyperplastic, dysplastic, metaplastic and neoplastic) could provide critical insights. Such validation would extend the applicability of these methods, potentially revolutionizing routine histopathology.
The widespread use of Ki-67 in clinical settings is attributed to its low cost, ease of use and minimal requirements for specialised equipment or advanced training. These characteristics align well with its integration into standard histopathological practices. Consequently, Ki-67 holds significant promise as a clinicopathological biomarker with potential applications in personalised therapy. Validating innovative methodologies to enhance its utility could make Ki-67 a cornerstone in tailoring cancer diagnostics and treatment strategies.
List of abbreviations
APC7, Anaphase-promoting complex subunit 7; APC/C-Cdh1, Anaphase-promoting complex Cdh1; CDE, Cell cycle dependent elements; CHR, Cell cycle genes homology regions; FUCCI, Fluorescent cell cycle indicator; Hklp2, Kinesin-like motor protein; LI, Labelling index; NET, Neuroendocrine tumours; NG, Nuclear gradient; NIFKh, Human nucleolar protein; NORs, Nucleolar organiser regions.
Acknowledgments
We thank CAPES, CNPQ and FAPERJ for the fellowships of the authors.
Conflicts of interest
The authors declare no conflicts of interest.
Funding
The authors have not received any kind of funding for the research and writing of this paper.
Consent for publication
All the authors of the article agreed to be published in the journal.
Author contributions
All authors contributed to the conception of the study. TANSD wrote this paper, and AKFS, NSCO, KSGC, CBPCS, HCC and ARS reviewed the paper. All authors had access to the study data and reviewed and approved the final manuscript.
References
1. Loftus LV, Amend SR, and Pienta KJ (2022) Interplay between cell death and cell proliferation reveals new strategies for cancer therapy Int J Mol Sci 23(9) 4723 https://doi.org/10.3390/ijms23094723 PMID: 35563113 PMCID: 9105727
2. Saint-Jacques N, Brown PE, and Purcell J, et al (2023) The nova scotia community cancer matrix: a geospatial tool to support cancer prevention Soc Sci Med 330 116038 https://doi.org/10.1016/j.socscimed.2023.116038 PMID: 37390806
3. Swanson K, Wu E, and Zhang A, et al (2023) From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment Cell 186(8) 1772–1791 https://doi.org/10.1016/j.cell.2023.01.035 PMID: 36905928
4. Edsjö A, Holmquist L, and Geoerger B (2023) Precision cancer medicine: Concepts, current practice, and future developments J Intern Med 294(4) 455–481 https://doi.org/10.1111/joim.13709 PMID: 37641393
5. Compton CC, Robb JA, and Anderson MW, et al Preanalytics and precision pathology: pathology practices to ensure molecular integrity of cancer patient biospecimens for precision medicine Arch Pathol Lab Med 143(11) 1346–1363 PMID: 31329478
6. Miller I, Min M, and Yang C, et al (2018) Ki67 is a graded rather than a binary marker of proliferation versus quiescence Cell Rep 24(5) 1105–1112.e5 https://doi.org/10.1016/j.celrep.2018.06.110 PMID: 30067968 PMCID: 6108547
7. Ali SA, Kadry MO, and Hammam O, et al (2022) Ki-67 pulmonary immunoreactivity in silver nanoparticles toxicity: Size-rate dependent genotoxic impact Toxicol Rep 9 1813–1822 https://doi.org/10.1016/j.toxrep.2022.09.011 PMID: 36518381 PMCID: 9742976
8. Miguel AFP, Poletto DAG, and Embaló B, et al (2023) Association between epithelial-mesenchymal transition markers, proliferative index, and oral epithelial dysplasia: an immunohistochemical study Oral Surg Oral Med Oral Pathol Oral Radiol 135(6) 904–913 https://doi.org/10.1016/j.oooo.2023.03.005 PMID: 37248101
9. Faragalla H, Plotkin A, and Barnes P, et al (2023) Ki67 in breast cancer assay: an ad hoc testing recommendation from the Canadian Association of Pathologists task force Curr Oncol 30(3) 3079–3090 https://doi.org/10.3390/curroncol30030233 PMID: 36975446 PMCID: 10047249
10. Razmi M, Tajik F, and Hashemi F (2024) The prognostic importance of Ki-67 in gastrointestinal carcinomas: a meta-analysis and multi-omics approach J Gastrointest Cancer 55(2) 599–624 https://doi.org/10.1007/s12029-024-01022-w PMID: 38411875
11. Ding L, Song L, and Zhao W et al (2020) Predictive value of p16INK4a, Ki-67 and ProExC immuno-qualitative features in LSIL progression into HSIL EExp Ther Med 19(4) 2457–2466
12. Li SD, Ba YP, and Zhang PW (2019) Prognosis factors analysis of 56 cases of non-Hodgkin's lymphoma originating from the nasopharynx Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 33(7) 651–653 PMID: 31327206
13. Menon SS, Guruvayoorappan C, and Sakthivel KM, et al (2019) Ki-67 protein as a tumour proliferation marker Clin Chim Acta 491 39–45 https://doi.org/10.1016/j.cca.2019.01.011 PMID: 30653951
14. Travaglino A, Raffone A, and Catena U, et al (2021) Ki67 as a prognostic marker in uterine leiomyosarcoma: a quantitative systematic review Eur J Obstet Gynecol Reprod Biol 266 119–124 https://doi.org/10.1016/j.ejogrb.2021.09.026 PMID: 34624740
15. Vlajnic T, Brunner P, and Eppenberger-Castori S, et al (2021) High inter- and intratumoral variability of Ki67 labeling index in newly diagnosed prostate cancer with high Gleason scores Pathobiology 89(2) 74–80 https://doi.org/10.1159/000519007 PMID: 34555829 PMCID: 9153326
16. Tao Z, Xue R, and Wei Z, et al (2023) The assessment of Ki-67 for prognosis of gastroenteropancreatic neuroendocrine neoplasm patients: a systematic review and meta-analysis Transl Cancer Res 12(8) 1980–1991 https://doi.org/10.21037/tcr-23-248 PMID: 37701110 PMCID: 10493787
17. Dias EP, Oliveira NSC, and Serra-Campos AO, et al (2021) A novel evaluation method for Ki-67 immunostaining in paraffin-embedded tissues Virchows Arch 479(1) 121–131 https://doi.org/10.1007/s00428-020-03010-4 PMID: 33464376
18. Booth DG, and Earnshaw WC (2017) Ki-67 and the chromosome periphery compartment in mitosis Trends Cell Biol 27(12) 906–916 https://doi.org/10.1016/j.tcb.2017.08.001 PMID: 28838621
19. Fonatsch C, Duchrow M, and Rieder H, et al (1991) Assignment of the human Ki-67 gene (MK167) to 10q25-qter Genomics 11(2) 476–477 https://doi.org/10.1016/0888-7543(91)90163-9 PMID: 1769665
20. Schmidt MH, Broll R, and Bruch HP, et al Proliferation marker pKi-67 occurs in different isoforms with various cellular effects J Cell Biochem 91(6) 1280–1292 PMID: 15048881
21. Chierico L, Rizzello L, and Guan L, et al (2017) The role of the two splice variants and extranuclear pathway on Ki-67 regulation innon-cancer and cancer cells PLoS One 12(2) e0171815 https://doi.org/10.1371/journal.pone.0171815 PMCID: 5302784
22. Uxa S, Castillo-Binder P, and Kohler R, et al (2021) Ki-67 gene expression Cell Death Differ 28(12) 3357–3370 https://doi.org/10.1038/s41418-021-00823-x PMID: 34183782 PMCID: 8629999
23. Sobecki M, Mrouj K, and Colinge J, et al (2017) Cell-cycle regulation accounts for variability in Ki-67 expression levels Cancer Res 77(10) 2722–2734 https://doi.org/10.1158/0008-5472.CAN-16-0707 PMID: 28283655
24. Müller GA, Asthana A, and Rubin SM (2022) Structure and function of MuvB complexes Oncogene 41(21) 2909–2919 https://doi.org/10.1038/s41388-022-02321-x PMID: 35468940 PMCID: 9201786
25. Kaulich M, Link VM, and Lapek JD, et al (2021) A Cdk4/6-dependent phosphorylation gradient regulates the early to late G1 phase transition Sci Rep 11(1) 14736 https://doi.org/10.1038/s41598-021-94200-w PMID: 34282211 PMCID: 8290049
26. Pattschull G, Walz S, and Gründl M, et al (2019) The Myb-MuvB complex is required for YAP-dependent transcription of mitotic genes Cell Rep 27(12) 3533–3546.e7 https://doi.org/10.1016/j.celrep.2019.05.071 PMID: 31216474
27. Andrés-Sánchez N, Fisher D, and Krasinska L (2022) Physiological functions and roles in cancer of the proliferation marker Ki-67 J Cell Sci 135(11) jcs258932 https://doi.org/10.1242/jcs.258932 PMID: 35674256
28. Guillaud P, du Manoir S, and Seigneurin D (1989) Quantification and topographical description of Ki-67 antibody labelling during the cell cycle of normal fibroblastic (MRC-5) and mammary tumour cell lines (MCF-7) Anal Cell Pathol 1(1) 25–39 PMID: 2488698
29. du Manoir S, Guillaud P, and Camus E, et al (1991) Ki-67 labeling in postmitotic cells defines different Ki-67 pathways within the 2c compartment Cytometry 12(5) 455–463 https://doi.org/10.1002/cyto.990120511 PMID: 1935459
30. van Dierendonck JH, Keijzer R, and van de Velde CJ, et al (1989) Nuclear distribution of the Ki-67 antigen during the cell cycle: comparison with growth fraction in human breast cancer cells Cancer Res 49(11) 2999–3006 PMID: 2720660
31. Sobecki M, Mrouj K, and Camasses A, et al (2016) The cell proliferation antigen Ki-67 organises heterochromatin Elife 5 e13722 https://doi.org/10.7554/eLife.13722 PMID: 26949251 PMCID: 4841783
32. Gerdes J, Li L, and Schlueter C, et al (1991) Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67 Am J Pathol 138(4) 867–873 PMID: 2012175 PMCID: 1886092
33. Ferguson CJ, Urso O, and Bodrug T, et al (2022) APC7 mediates ubiquitin signaling in constitutive heterochromatin in the developing mammalian brain Mol Cell 82(1) 90–105.e13 https://doi.org/10.1016/j.molcel.2021.11.031 PMCID: 8741739
34. MacCallum DE and Hall PA (1999) Biochemical characterization of pKi67 with the identification of a mitotic-specific form associated with hyperphosphorylation and altered DNA binding Exp Cell Res 252(1) 186–198 https://doi.org/10.1006/excr.1999.4600 PMID: 10502411
35. Booth DG, Takagi M, and Sanchez-Pulido L, et al (2014) Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery Elife 3 e01641 https://doi.org/10.7554/eLife.01641 PMID: 24867636 PMCID: 4032110
36. Matheson TD and Kaufman PD (2017) The p150N domain of chromatin assembly factor-1 regulates Ki-67 accumulation on the mitotic perichromosomal layer Mol Biol Cell 28(1) 21–29 https://doi.org/10.1091/mbc.e16-09-0659 PMCID: 5221625
37. Stamatiou K, Huguet F, and Serapinas LV, et al (2024) Ki-67 is necessary during DNA replication for fork protection and genome stability Genome Biol 25(1) 105 https://doi.org/10.1186/s13059-024-03243-5 PMID: 38649976 PMCID: 11034166
38. Sasaki K, Matsumura K, and Murakami T, et al (1990) Intranuclear localization of the Ki-67 reactive antigen in HeLa cells flow cytometric analysis Biol Cell 68(2) 129–132 https://doi.org/10.1016/0248-4900(90)90297-G PMID: 2192767
39. Saiwaki T, Kotera I, and Sasaki M, et al (2005) In vivo dynamics and kinetics of pKi-67: transition from a mobile to an immobile form at the onset of anaphase Exp Cell Res 308(1) 123–134 https://doi.org/10.1016/j.yexcr.2005.04.010 PMID: 15896774
40. Trerè D, Farabegoli F, and Cancellieri A, et al (1991) AgNOR area in interphase nuclei of human tumours correlates with the proliferative activity evaluated by bromodeoxyuridine labelling and Ki-67 immunostaining J Pathol 165(1) 53–59 https://doi.org/10.1002/path.1711650109 PMID: 1955936
41. Starborg M, Gell K, and Brundell E, et al (1996) The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression J Cell Sci 109(Pt 1) 143–153 https://doi.org/10.1242/jcs.109.1.143 PMID: 8834799
42. Rahmanzadeh R, Hüttmann G, and Gerdes J, et al (2007) Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis Cell Prolif 40(3) 422–430 https://doi.org/10.1111/j.1365-2184.2007.00433.x PMID: 17531085 PMCID: 6496591
43. Cuylen S, Blaukopf C, and Politi AZ, et al Ki-67 acts as a biological surfactant to disperse mitotic chromosomes Nature 535(7611) 308–312 PMID: 27362226 PMCID: 4947524
44. Stamatiou K, Chmielewska A, and Ohta S, et al (2023) CCDC86 is a novel Ki-67-interacting protein important for cell division J Cell Sci 136(2) jcs260391 https://doi.org/10.1242/jcs.260391 PMID: 36695333 PMCID: 10022746
45. Caragine CM, Haley SC, and Zidovska A (2019) Nucleolar dynamics and interactions with nucleoplasm in living cells Elife 8 e47533 https://doi.org/10.7554/eLife.47533 PMID: 31769409 PMCID: 6879204
46. Li W, Lu N, and Chen C, et al (2023) Identifying the optimal cutoff point of Ki-67 in breast cancer: a single-center experience J Int Med Res 51(8) 3000605231195468 https://doi.org/10.1177/03000605231195468 PMID: 37652458 PMCID: 10478558
47. Ai D, Turashvili G, and Gjeorgjievski SG, et al (2024) Subspecialized breast pathologists have suboptimal interobserver agreement in Ki-67 evaluation using 20% as the cutoff Breast Cancer Res Treat Apr 204(2) 415–422 https://doi.org/10.1007/s10549-023-07197-3 PMID: 38157098
48. Mungle T, Tewary S, and Arun I, et al (2017) Automated characterization and counting of Ki-67 protein for breast cancer prognosis: a quantitative immunohistochemistry approach Comput Methods Programs Biomed 139 149–161 https://doi.org/10.1016/j.cmpb.2016.11.002 PMID: 28187885
49. Dawe M, Shi W, and Liu TY, et al Reliability and variability of Ki-67 digital image analysis methods for clinical diagnostics in breast cancer Lab Invest 104(5) 100341 PMID: 38280634
50. Witkiewicz AK, Kumarasamy V, and Sanidas, et al (2022) Cancer cell cycle dystopia: heterogeneity, plasticity, and therapy Trends Cancer 8(9) 711–725 https://doi.org/10.1016/j.trecan.2022.04.006 PMID: 35599231 PMCID: 9388619
51. Maleki EH, Bahrami AR, and Matin MM (2023) Cancer cell cycle heterogeneity as a critical determinant of therapeutic resistance Genes Dis 11(1) 189–204 https://doi.org/10.1016/j.gendis.2022.11.025 PMID: 37588236 PMCID: 10425754
52. Faviana P, Boldrini L, and Gentile C, et al (2022) Proposal for a new diagnostic histopathological approach in the evaluation of Ki-67 in GEP-NETs Diagnostics 12(8) 1960 https://doi.org/10.3390/diagnostics12081960 PMID: 36010311 PMCID: 9407142
53. Fernezlian SM, Baldavira CM, and Souza MLF, et al (2023) A semi-automated microscopic image analysis method for scoring Ki-67 nuclear immunostaining Braz J Med Biol Res 56 e12922 https://doi.org/10.1590/1414-431x2023e12922 PMID: 37970922 PMCID: 10644968
54. Guo X and Chen L (2024) From G1 to M: a comparative study of methods for identifying cell cycle phases Brief Bioinform 25(2) bbad517 https://doi.org/10.1093/bib/bbad517 PMID: 38261342 PMCID: 10805071
55. Yang HW (2024) Investigating heterogeneous cell-cycle progression using single-cell imaging approaches Methods Mol Biol 2740 263–273 https://doi.org/10.1007/978-1-0716-3557-5_16 PMID: 38393481
56. Chen Y, Zhang H, and Fan Y, et al (2024) Multiplex cyclic fluorescent immunohistochemistry J Vis Exp 203 [doi: 10.3791/66136] https://doi.org/10.3791/66136]
57. Baumhoer D, Hench J, and Amary F (2024) Recent advances in molecular profiling of bone and soft tissue tumors Skeletal Radiol 53(9) 1925–1936 https://doi.org/10.1007/s00256-024-04584-9 PMID: 38231260 PMCID: 11303483
58. Pochechueva TV, Schwenzer N, and Kohl T, et al (2024) 3D super-resolution nuclear Q-FISH imaging reveals cell-cycle-related telomere changes Int J Mol Sci 25(6) 3183 https://doi.org/10.3390/ijms25063183 PMID: 38542157 PMCID: 10970233