Low prostaglandin-endoperoxide synthase-2 gene expression in colorectal carcinomas may predict poorer survival
Uchenna Simon Ezenkwa1,2, Sebastian Anebuokhae Omenai3, Oluwadamilare Iyapo4, Chinedu Anthony Ezekekwu5, Adesoji E Adetona6, Chima Uzoma Akunwata7, Ayotunde Oladunmi Ale8 and Henry Okwuchukwu Ebili6
1Department of histopathology, Federal University of health Sciences Azare, Azare 751101, Bauchi, Nigeria
2Department of Pathology, Federal Medical Centre Azare, Azare 751101, Bauchi, Nigeria
3Department of Anatomical Pathology, Edo State University Uzairue, Auchi 312002, Edo, Nigeria
4Department of Pathology, Federal Medical Centre, Ebute Metta, Lagos 101212, Nigeria
5Bristol haematology and Oncology Centre, University hospital, Bristol and Weston NhS Trust, Bristol BS28ED, UK
6Department of Morbid Anatomy & histopathology, Olabisi Onabanjo University, Ago-Iwoye 121101, Ogun State, Nigeria
7Department of haematology, University College hospital Ibadan, Ibadan 200221, Oyo, Nigeria
8Department of Medicine, Olabisi Onabanjo University/University Teaching hospital, Sagamu 121101, Ogun, Nigeria
ahttps://orcid.org/0000-0002-7022-8268
bhttps://orcid.org/0000-0002-1841-1487
chttp://orcid.org/0000-0003-1102-094X
dhttps://orcid.org/0000-0003-3703-2316
ehttps://orcid.org/0000-0002-7548-5087
fhttps://orcid.org/0000-ooo3-1779-7311
ghttps://orcid.org/0000-0002-2135-1796
Abstract
Introduction: Prostaglandin-endoperoxide synthase-2 (ptgs2), otherwise called Cyclooxygenase 2, is overexpressed in colorectal carcinoma (CRC) compared to normal tissues. however, the impact of differential expression among ptgs2-positive tumours on CRC prognosis has not been well investigated. By sub-stratifying positive tumour expression, this study determined its potential influence on patients’ outcomes.
Methods: The Cancer Genome Atlas database was explored to determine CRC cases with RNA-Sequence (RNA-Seq) transcript data and matched clinicopathological data alongside gene copy number variation and methylation status. Descriptive, chi-square, Fisher exact, Linear-by-Linear associations, logistic and Kaplan-Meier statistics were used to determine proportions, associations, predictors and survival between ptgs2 and tumour parameters using Statistical Package for Social Sciences version 20. Two-tailed p-value <0.05 was accepted as statistically significant.
Results: There were 534 CRC classified predominantly as adenocarcinoma not otherwise specified (86.3%) and mucinous carcinoma (12.4) histologically included in this study. Marker (ptgs2) expression ranged from 0.02 FPKM-131.89 FPKM, (Median 1.4 FPKM). The majority of the cases (53.4%) were diagnosed at an early stage and showed high ptgs2 RNA-Sequence (RNA-seq) expression in 51.5% (275/534). Significant associations were seen between ptgs2 expression and histological subtype (p < 0.001), lymphovascular invasion (p = 0.013), pN2 stage (> 6 positive lymph nodes) (p = 0.011) and American Joint Committee on Cancer Staging stage (p = 0.028), and these all had lower ptgs2 expression. On regression analysis, histological differentiation emerged as a predictor of ptgs2 expression (Odds ratio 2.749, 95% confidence interval 1.479–5.108, p < 0.001). Also, gene methylation was associated with reduced ptgs2 expression. Overall survival was significantly inferior among individuals with low ptgs2 tumours (p = 0.018) while that for disease-free survival was non-significant (p = 0.327).
Conclusion: CRCs with low ptgs2 transcripts are associated with poorer survival. This finding suggests a need for closer follow up and tailored adjuvant therapy for these patients.
Keywords: prostaglandin-endoperoxide synthase-2, cyclooxygenase-2, colorectal carcinoma, TCGA, prognosis, survival
Correspondence to: Uchenna Simon Ezenkwa
Email: uchennaezenkwa@gmail.com
Published: 06/12/2024
Received: 13/03/2024
Publication costs for this article were supported by ecancer (UK Charity number 1176307).
Copyright: © the authors; licensee ecancermedicalscience. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Colorectal carcinoma (CRC) is the most common malignancy of the gastrointestinal tract [1]. Global estimates suggest that there were about 1.9 million new CRC cases and 904,000 deaths in 2022, ranking third in terms of incidence but second in terms of mortality from all cancers [2]. This high burden is disproportionately distributed between developed and developing countries, the former having higher incidence while the latter has higher burden of mortality rates, respectively [1, 3]. Alcohol consumption, tobacco smoking, consumption of red or processed meat and increase in body fatness raise the risk of developing the disease while calcium supplements, whole grains, fiber and dairy products consumption, as well as physical activity, are considered protective, particularly for colon cancer [4]. The increasing adoption of cancer-promoting lifestyles and less protective ones, especially among low- and middle-income countries shows that the burden of the disease is bound to increase [2]. To mitigate this impact, there is a need to refine our understanding of the disease process for a more effective preventive or treatment approach.
Carcinogenesis is a multistage process characterised by cumulative genetic alterations in cellular homeostatic replicating pathways [5]. In the colorectum, for example, it has been shown that gain-of-function changes in genes promoting or loss-of-function in those suppressing cell proliferation are fundamental to tumourigenesis [6]. Usually, the transformation of normal epithelium to carcinoma begins with the dysregulation of the canonical Wnt pathway mediated by a mutated APC gene and gives rise to a dysplastic mucosa (adenoma) [7]. Subsequent accumulation of other genetic defects such as SMAD4/2, hTERT and TP53 lead to advanced adenoma and carcinoma. Other established pathways include loss of DNA mismatch repair capability, chromosomal instability (resulting from aneuploidy, deletions, insertions, amplifications or loss of heterozygosity), microsatellite instability, serrated adenoma pathway (activating V600E mutation in BRAF gene) and DNA methylation [8].
Recent studies suggest that synergistic pathways involving genes regulating the tumour microenvironment, histone acetylation and inflammation can also influence the carcinoma process. Perturbations in COL11A1 and hDAC9 are believed to induce disturbances in some signaling pathways such as the RTK-RAS-PI3K, Wnt, TGF-β and TP53 in the colon, thereby influencing cancer development [9, 10]. Toll-like receptor signaling in the colon has also been implicated in tumourigenesis through the induction of chronic inflammation within the gut [11]. Indeed, signature genes responsible for the progression to CRC in patients with ulcerative colitis have been proposed, including WFDC2, TTLL12, ThRA, EPhB3 [12], ARID1A, FBXW7, KRAS, RNF, APC, P53 and SMAD4 genes [13]. Thus, these indicate a putative role of inflammatory mediators in carcinogenesis [12].
One such inflammatory mediator is prostaglandin E2, a byproduct of cyclooxygenase-2 (COX-2) enzyme action on arachidonic acids [14]. In 1994, Eberhart et al [15] described a differential overexpression of COX-2 mRNA in CRC and adenomatous polyps compared to normal tissues and proposed that COX-2 could be a control target for CRC [15]. Subsequent studies investigating this observation have reached varying conclusions, a situation largely due to differing methods of detecting and/or scoring the marker expression [16]. For example, Sheehan et al [17] found heterogenous tumour staining with COX 2 antibodies and further showed that more low-grade tumours with COX-2 positivity had better overall survival than high-grade tumours [17]. Likewise, Ogino et al [18] using immunohistochemistry, showed that COX-2 positive patients had inferior colon cancer-specific mortality even after adjusting for the stage and clinical characteristics of the patients [18]. On the other hand, Zahedi et al [19] also observed that COX-2 expression by tumour cells had no influence on tumour invasiveness or patient outcome, even though tumour tissues had higher COX-2 expression compared to normal tissues. As such, they opined that iCOX-2 plays a role in initiating the carcinogenic process but not its progression [19].
The varying mutational landscapes illustrated above show that CRC is a heterogenous disease, and although much is known about it, its 5-year survival is still below optimum despite therapies informed by these advances. There is therefore a need to interrogate further what is known in order to gain more insights that can help improve outcomes. Most studies investigating the role of ptgs2 (COX-2) in CRC have focused on differential expression between normal and malignant tissues or between marker-expressing and non-expressing tumours. This presupposes that tumours expressing ptgs2 behave alike irrespective of the degree to which each tumour expresses the marker. however, while evidence supports the hypothesis that COX-2 expression among COX-2-expressing tumours is not uniform [17], the potential influence of this variation on the prognosis and clinicopathological features of CRC has not been sufficiently investigated. The present study relied on ptgs2 transcripts data from The Cancer Genome Atlas (TCGA) database, to determine the association between high and low ptgs2 expression and patient survival as a primary endpoint, and by extension its relationship with clinicopathological factors.
Materials and methods
RNA-Seq data of CRCs stored as Colorectal Adenocarcinoma (TCGA, PanCancer Atlas) was downloaded from TCGA repository at cBioportal [20]. Additionally, data on copy number variation (CNV) and methylation status of the genes were also retrieved. The procedure for extracting such data is well explained by Gao et al [21]. Matched patient clinical and tumour histopathological information such as gender, age at diagnosis, tumour location within the large bowel (colon or rectum), tumour histological subtype, tumour depth (pT), tumour lymph node involvement, number of positive lymph nodes (pN), tumour metastasis, lymphovascular invasion, vascular permeation and tumour staging according to the American Joint Committee on Cancer Staging, overall and disease-free survival (DFS) in months were extracted too. having a documented ptgs2 RNA gene transcripts were mandatory for inclusion into the study while cases without documented ptgs2 transcripts were excluded. The data used in this study was derived from a published source with complete participant de-identification, and therefore does not require ethical review or approval.
Data analysis
Statistical analysis was applied to the obtained data using the Statistical Package for Social Sciences version 20. Association between ptgs2 expression level and overall survival was the primary endpoint while DFS and association with overall American Joint Committee on Cancer Staging (AJCC) tumour stage were secondary endpoints. Categorical variables were summarized as proportions while continuous variables were described as median using descriptive statistical tool. The association between ptgs2 overexpression and the clinicopathological parameters was tested using Pearson’s chi-square, Fisher exact test or Linear-by-Linear associations. Also, the association between ptgs2 expression and ptgs2 CNV was evaluated using linear-by-linear association statistics, while the relationship between ptgs2 and methylation status was determined through Spearman correlation analysis. Binomial logistic regression was used to test for predictors of ptgs2 expression while Kaplan-Meier statistics was applied to the data to test for association between ptgs2 and overall and DFS. All p-value were determined at a two-tailed significance level of <0.05. Outputs were presented as text, tables and figures.
Results
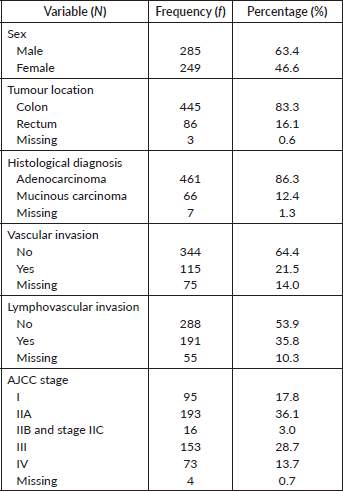
There were 629 entries for CRC on the dataset out of which, 95 (15.1%) had no ptgs2 data and were excluded from the study. Five hundred and thirty-four (84.9%) cases comprising of 285 males and 249 females with a median age of 68 years (age range 31–90 years) were therefore included in the study. Table 1 shows the patient and tumour pathological characteristics. The tumours were broadly categorised as adenocarcinoma not otherwise specified (86.3%) and mucinous carcinoma (12.4%). Most of the tumours were located within the colon (83%) and the majority were seen at an early stage (AJCC I-IIA).
Table 1. Clinicopathological features of the study population.
PTGS2 gene expression pattern
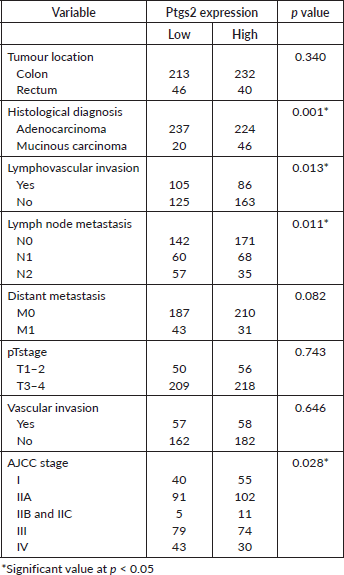
The dichotomised ptgs2 expression profile showed RNA overexpression in 51.5% (275) of the CRCs. The median expression was 1.4 FPKM ranging from 0.02 to 131.89 FPKM. Table 2 shows associations between ptgs2 expression and the tumour characteristics. Mucinous carcinomas were more likely to express high ptgs2 compared to adenocarcinomas (p < 0.001). Other observed significant associations were between ptgs2 expression and lymphovascular invasion (p = 0.013), lymph node metastasis (p = 0.011) and tumour stage (p = 0.028). The majority of tumours with lymphovascular invasion by tumour cells had low ptgs2 expression (105/191; 55%). In contrast, tumours without lymphovascular invasion had higher ptgs2 (163/288, 56.6%). Likewise, CRC cases having early AJCC tumour staging (I-IIA) showed higher ptgs2 compared to late-stage tumours (AJCC IIB – IV). Tests of association between ptgs2 and vascular, pM and pT staging did not reach statistical significance (Table 2).
Logistic regression for predictive markers of ptgs2 expression
The result of the statistical test for predictors of ptgs2 expression in CRC using binomial logistic regression is as shown in Table 3. Only the histological subtype significantly predicted COX-2 gene expression (p < 0.001). AJCC tumour stage, lymphovascular invasion and lymph node metastasis status did not reach acceptable level of statistical significance (p > 0.05).
Table 2. Association between ptgs2 expression pattern and colorectal cancer pathological characteristics.
CNV, methylation status and ptgs2 expression
There was available CNV data for 524 tumour tissues. COX-2 gene deletions and amplifications were seen in 50 (9.54%) and 117 (22.33%) cases, respectively, while wild-type variants were 357 (68.13%). The association between CNV and ptgs2 expression was statistically non-significant.
The correlation between ptgs2 and overall methylation status was both inverse and significant (r = −0.289; p < 0.001). Methylation analysis involved several loci, with some of these loci identified in as few as 156 tumour tissue samples, while others were detected in up to 516 cases. Only loci expression data present for up to a minimum of 300 patients was included in this analysis. Most of the loci had an inverse relationship with ptgs2 expression. These include COX2_cg00690431 (r = −0.098; p = 0.076), COX2_cg04881125 (r = −0.081; p = 0.142), COX2_cg07422329 (r = −0.168; p = 0.002), COX2_cg08482694 (r = −0.102; p = 0.021), COX2_cg10180406 (r = −0.201; p < 0.001), COX2_cg13986130 (r = −0.067; p = 0.126), COX2_cg16101346 (r = −0.311; p < 0.001), COX2_cg18335243 (r = −0.090; p = 0.041), COX2_cg22365834 (r = −0.058; p = 0.310), COX2_cg23070111 (r = −0.126; p = 0.022), COX2_cg24887140 (r = 0.173; p = 0.002), COX2_cg25147026 (r = −0.331; p < 0.001), COX2_cg25837803 (r = −0.235; p < 0.001) and COX2_cg26564040 (r = −0.245; p < 0.001). Two loci showed positive correlations and included COX2_cg07422329 (r = 0.388; p < 0.001) and COX2_cg17419623 (r = 0.070; p = 0.205).
Segregating between low and high ptgs2 expression, there was a statistically significant difference in the median methylation status (expression) between low and high ptgs2 expressing tumours. Low ptgs2 CRC had higher gene methylation compared to high ptgs2 CRC (Figure 1). Sustained significant differential methylation between low and high ptgs2-expressing tumours was observed at loci COX2_cg07422329 (p < 0.001), COX2_cg08482694 (p = 0.036), COX2_cg16101346 (p < 0.001), COX2_cg18335243 (p = 0.028), COX2_cg24887140 (p = 0.003) and COX2_cg25147026 (p < 0.001).
Table 3. Binomial logistic regression assessing predictors of ptgs2 expression in CRC.
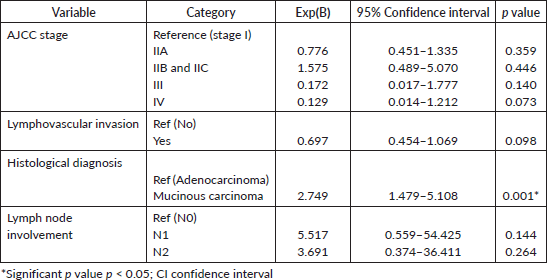
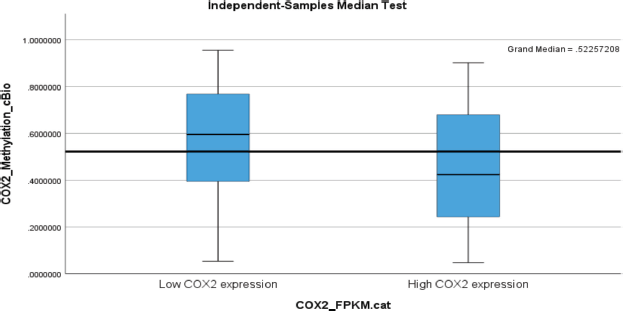
Figure 1. A box plot showing higher PTGS2 (COX-2) methylation status in low PTGS2-expressing colorectal carcinoma with its median value being above the overall median of 0.52.
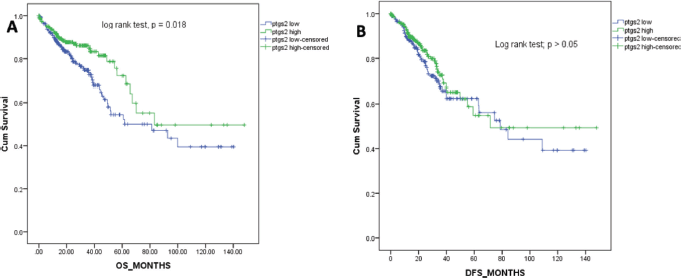
Figure 2. Kaplan-Meier survival curves for the overall survival (a) and DFS (b) in months.
Ptgs2 expression and survival outcomes
The overall survival for the patients ranged from 0 to 148 months with a median of 21.1 months. Figure 2 shows the Kaplan-Meier survival curve for the overall survival of the patients included in this study. Patients with higher ptgs2 expression had better survival outcomes than those with low ptgs2. The Log Rank (Mantel-Cox) test for survival outcome was statistically significant (Figure 2A). The median DFS for this population was 20 (0–148) months. Low ptgs2 tumours had poorer DFS although there was no significant difference in the DFS between the two groups as shown by the Log Rank (Mantel-Cox) test (p = 0.327, Figure 2B).
Discussion
The findings of this study showed that CRC tumours expressing ptgs2 do not all behave similarly. In addition, those with lower marker expression had significantly poorer overall survival. Also, although DFS was not significantly different between ptgs2-low and high tumours, ptgs2-low tumour patients had inferior DFS compared to ptgs2-high tumour patients. Few authors have documented similar findings in colorectal and breast cancers previously [22, 23]. Kim et al [22] showed that CRC patients with elevated COX-2 expression experienced favourable recurrence-free survival compared to COX-2-low tumours with overexpressing cancers having the late onset of recurrence [22]. Similarly, recent meta-analysis data have demonstrated better overall and disease-specific survival in high COX-2-expressing CRC [24, 25]. In contrast, Wu and Sun [26], Kosumi et al [27] and Mima et al [28] found higher mortality among high COX-2 expressing tumour patients in their respective studies. Notably, these studies classified both low and negative COX-2-expressing tumours in one category, [26–28] whereas the present study focused only on tumours with ptgs2 transcripts. More studies are however needed to further test these observations and determine other meaningful ways of interpreting the biomarker in CRC.
Notwithstanding these observed differences, our result has implications for CRC research and care. Evidence shows that the use of anti-COX-2 agents improves disease-specific survival in CRC patients post-diagnosis [25]. We believe that the potential benefit of COX-2 inhibition would apply to both low- and high-ptgs2 tumours, although, patients with low ptgs2 tumours may require a closer follow-up and individualized adjuvant therapy to control their disease. This highlights a need to sub-categorise ptgs2 expression in CRC tumours such that low marker tumours are delineated from high marker tumours and also from those in which the marker is absent or undetected, and testing their prognostic relationships in future study designs.
Whilst lower CRC disease survival among low-ptgs2-tumours was unexpected, it nevertheless offers insights into the various ways by which the emerging role of the biomarker can be interrogated. First, the results showed that early-stage tumours, those without lymphovascular invasion and those having fewer numbers of lymph node metastasis, had higher ptgs2 expression than advanced tumours. This suggests that the biomarker may have been produced at a higher magnitude during tumor initiation, potentially playing a role in promoting the early stages of tumor development [16, 19]. There are literatures supporting this position. Ptgs2 is an inducible gene and was shown at its primary characterization to be induced by the mitogen Rous Sarcoma Virus with its product, PGE2, mediating tumour cell proliferation [29, 30]. Its (ptgs2) expression has been shown to promote, while its inhibition prevents colorectal adenomas [30, 31]. Williams et al [32] showed from their experiment through COX-2 inhibition, that COX-2 mediates transitions in the cell cycle by lifting G2/M blockade. Other researchers have also observed that COX-2 promotes cell proliferation in CRC by up-regulating micro-RNA miR-21, an antagonist of PGE2-degrading enzymes and tumour suppressor genes 15-PGDh and PDCD4 [33]. Likewise, COX-2 cross-talk with p53 led to inverse reciprocal regulation that reduced p53-mediated apoptosis in CRC in another study [34]. These observations therefore suggest a role for ptgs2 in initiating colorectal carcinogenesis [16].
Second, the differential expression of higher ptgs2 among early-stage tumours could support an alternative role as a tumour suppressor. From our result, the diminished ptgs2 expression seen with more invasive tumour characteristics may suggest that the marker expression is suppressed from its initial rate of production as the tumour advances. This loss-of-function could therefore lead to a more aggressive disease phenotype leading to worse overall survival among these patients. The ptgs2 methylation data also showed that the gene expression reduced in proportion to the methylation status, suggesting ptgs2 methylation as a regulatory mechanism in CRC. One mechanism by which ptgs2 could influence tumour suppression is through interaction with other mediators. Studies have shown that ptgs2 overexpression up regulates Kruppel-like factor-4 (KLF4), a tumour suppressor gene that mediates apoptosis in dividing cells but is lost in CRC [35, 36]. Thus, loss or downregulation of ptgs2 could potentially lead to dysregulation of KLF4 and similar mediators, leading to poor disease outcomes.
Another plausible explanation relates to synergistic interaction among ptgs2, tumour pathological factors and other molecular mediators. For example, tumour stage is an established independent prognostic factor in CRC [37] and likely played the most significant role in this study. Also, the interplay between ptgs2 and other well-known molecular markers of CRC prognosis has become a subject of recent investigation among researchers. Increased COX-2 is associated with increased matrix metalloproteinase-7 synthesis in colonic polyps, thus preparing the ground for invasiveness; post-diagnosis aspirin intake showed more benefit in tumours with coexisting high COX-2 and mutated PIK3C gene; COX-2-derived PGE2 induces PD-1 expression by CRC cells thereby evading host immune system; while inhibition of COX-2 re-sensitises resistant Epidermal Growth Factor Receptor (EGFR) receptors in CRC and increases tumour cell death by down-regulating miR-21 with elevated PDCD4 (a tumour suppressor) and 15-PGDh (a PGE2 degrading enzyme); to mention a few [38–43]. Therefore, the overall interpretation of the present study should be done without overlooking the roles these mediators and tumour histopathological characteristics could play in the disease progression and outcome.
Further on the histopathological variables, we found significantly elevated ptgs2 in mucinous carcinomas and this was sustained on logistic regression. Mucinous CRC are known to have high PIK3C, microsatellite instability (MSI) and BRAF mutations and are more susceptible to PD-1 inhibition [44]. They also tend to influence poorer survival although those with high MSI status show better outcomes [45, 46]. how this interacts to influence tumour progression needs to be elucidated. however, we think that the higher ptgs2 observed among mucinous carcinomas could be as a result of the stimulating effect of mucin on ptgs2 expression in the tumour microenvironment that results in the upregulation of these genes, further suggesting a role for inflammatory mediators such as COX-2 in tumour progression [47]. Therefore, higher ptgs2 levels in mucinous carcinoma, lower amounts in more invasive cancers, together with the influence of tumour staging gives us confidence that indeed, ptgs2 expression in this study had prognostic value.
There were several limitations to this study. First, the limited number of studies investigating the prognostic impact of the PTGS2 (COX-2) gene expression in marker-expressing colorectal cancers (CRCs) made it challenging to compare our findings with existing literature. This scarcity of data highlights the need for more comprehensive research in this area. Second, it was not feasible to determine whether patients had prior exposure to COX-2 inhibitors before surgery, a factor that could potentially influence PTGS2 expression and affect our observations. This limitation could be addressed in future studies by incorporating patient histories regarding COX-2 inhibitor use into a prospective study design. Overall, further research is necessary to explore the hypothesis that positive ptgs2 expression or its product, COX-2, in CRC should be further stratified to enhance prognostic benefit in patient management, as we suggest in our findings.
Conclusion
Sub-categorising CRC tumours expressing ptgs2 into low and high expressors showed a significant effect on overall survival in this study. This suggests that all marker-expressing tumours do not behave alike and thus, may need to be treated differently. While this could suggest novel approaches in CRC therapy, caution is rather advised in interpreting these findings without more studies.
Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.
Funding
The authors declare that no funding or other form of support was received for this study.
Consent to participate
Not required.
Consent to publish
Not required.
Ethics approval
The data used in this study are available on the public domain, hence no ethical approval was required.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by USE, hOE, SAO and AOA. The first draft of the manuscript was written by USE and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
References
1. Arnold M, Abnet CC, and Neale RE, et al (2020) Global burden of 5 major types of gastrointestinal cancer Gastroenterology [Internet] 159 335–349.e15 [https://linkinghub.elsevier.com/retrieve/pii/S0016508520304522] Date accessed: 12/10/24 https://doi.org/10.1053/j.gastro.2020.02.068 PMID: 32247694 PMCID: 8630546
2. Bray F, Laversanne M, and Sung H, et al (2024) Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin [Internet] 74 229–263 Date accessed: 01/07/24 https://doi.org/10.3322/caac.21834 PMID: 38572751
3. Goodarzi E, Beiranvand R, and Naemi H, et al (2019) Worldwide incidence and mortality of colorectal cancer and human development index (HDI): an ecological study WCR J 6 1–10
4. World Cancer Research Fund/American Institute for Cancer Research. Continuous Update Project Expert Report 2018 (2018) Diet, Nutrition, Physical Activity and Colorectal Cancer [dietandcancerreport.org]
5. Nunney L (2016) Commentary: the multistage model of carcinogenesis, Peto’s paradox and evolution Int J Epidemiol [Internet] 45 649–653 Date accessed: 12/10/24 https://doi.org/10.1093/ije/dyv201
6. Sommariva S, Caviglia G, and Piana M (2021) Gain and loss of function mutations in biological chemical reaction networks: a mathematical model with application to colorectal cancer cells J Math Biol [Internet]82 55 Date accessed: 12/10/24 https://doi.org/10.1007/s00285-021-01607-0 PMID: 33945019 PMCID: 8096774
7. Zhang L and Shay JW (2017) Multiple roles of APC and its therapeutic implications in colorectal cancer JNCI J Natl Cancer Inst [Internet] 109 djw332 [https://academic.oup.com/jnci/article/doi/10.1093/jnci/djw332/3113843] Date accessed: 01/10/24 https://doi.org/10.1093/jnci/djw332 PMID: 28423402 PMCID: 5963831
8. Nguyen LH, Goel A, and Chung DC (2020) Pathways of colorectal carcinogenesis Gastroenterology [Internet] 158 291–302 [https://linkinghub.elsevier.com/retrieve/pii/S0016508519414418] Date accessed: 01/10/24 https://doi.org/10.1053/j.gastro.2019.08.059
9. Patra R, Das NC, and Mukherjee S (2021) Exploring the differential expression and prognostic significance of the COL11A1 gene in human colorectal carcinoma: an integrated bioinformatics approach Front Genet [Internet] 12 608313 [https://www.frontiersin.org/articles/10.3389/fgene.2021.608313/full] Date accessed: 12/10/24 https://doi.org/10.3389/fgene.2021.608313 PMID: 33597969 PMCID: 7882494
10. Patra R, Chakraborty J, and Das NC, et al (2023) An integrated omics study on the role of HDAC9 gene in the oncogenic events of human gastrointestinal-tract associated cancers Hum Gene [Internet] 37 201189 [https://linkinghub.elsevier.com/retrieve/pii/S2773044123000487] Date accessed: 12/10/24 https://doi.org/10.1016/j.humgen.2023.201189
11. Mukherjee S, Patra R, and Behzadi P, et al (2023) Toll-like receptor-guided therapeutic intervention of human cancers: molecular and immunological perspectives Front Immunol [Internet] 14 1244345 [https://www.frontiersin.org/articles/10.3389/fimmu.2023.1244345/full] Date accessed: 12/10/24 https://doi.org/10.3389/fimmu.2023.1244345
12. Patra R, Dey AK, and Mukherjee S (2023) Identification of genes critical for inducing ulcerative colitis and exploring their tumorigenic potential in human colorectal carcinoma PLOS One [Internet] 18 e0289064 Date accessed: 12/10/24 https://doi.org/10.1371/journal.pone.0289064
13. Zhou RW, Harpaz N, and Itzkowitz SH, et al (2023) Molecular mechanisms in colitis-associated colorectal cancer Oncogenesis [Internet] 12 48 Date accessed: 12/10/24 https://doi.org/10.1038/s41389-023-00492-0
14. Wang B, Wu L, and Chen J, et al (2021) Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets Signal Transduct Target Ther [Internet] 6 94 Date accessed: 13/10/24 https://doi.org/10.1038/s41392-020-00443-w
15. Eberhart CE, Coffey RJ, and Radhika A, et al (1994) Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas Gastroenterology [Internet] 107 1183–1188 Date accessed: 01/10/24 https://doi.org/10.1016/0016-5085(94)90246-1
16. Negi RR, Rana SV, and Gupta V, et al (2019) Over-expression of cyclooxygenase-2 in colorectal cancer patients Asian Pac J Cancer Prev [Internet] 20 1675–1681 [http://journal.waocp.org/article_88244.html] Date accessed: 01/10/24 https://doi.org/10.31557/APJCP.2019.20.6.1675
17. Sheehan KM, Sheahan K, and O’Donoghue DP, et al (1999) The relationship between cyclooxygenase-2 expression and colorectal cancer JAMA [Internet] 282 1254 Date accessed: 01/10/24 https://doi.org/10.1001/jama.282.13.1254
18. Ogino S, Kirkner GJ, and Nosho K, et al (2008) Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer Clin Cancer Res [Internet] 14 8221–8227 [https://aacrjournals.org/clincancerres/article/14/24/8221/73328/Cyclooxygenase-2-Expression-Is-an-Independent] Date accessed: 01/10/24 https://doi.org/10.1158/1078-0432.CCR-08-1841
19. Zahedi T, Colagar AH, and Mahmoodzadeh H (2021) PTGS2 over-expression: a colorectal carcinoma initiator not an invasive factor Rep Biochem Mol Biol 9 442–451 https://doi.org/10.52547/rbmb.9.4.442 PMID: 33969138 PMCID: 8068447
20. Cerami E, Gao J, and Dogrusoz U, et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data Cancer Discov [Internet] 2 401–404 [https://aacrjournals.org/cancerdiscovery/article/2/5/401/3246/The-cBio-Cancer-Genomics-Portal-An-Open-Platform] Date accessed: 01/10/24 https://doi.org/10.1158/2159-8290.CD-12-0095
21. Gao J, Aksoy BA, and Dogrusoz U, et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal Sci Signal [Internet] 6 pl1 Date accessed: 01/10/24 https://doi.org/10.1126/scisignal.2004088
22. Kim SH, Ahn BK, and Paik SS, et al (2018) Cyclooxygenase-2 expression is a predictive marker for late recurrence in colorectal cancer Gastroenterol Res Pract [Internet] 2018 1–8 [https://www.hindawi.com/journals/grp/2018/7968149/] Date accessed: 01/10/24 https://doi.org/10.1155/2018/7968149
23. Saindane M, Rallabandi HR, and Park KS, et al (2020) Prognostic significance of prostaglandin-endoperoxide synthase-2 expressions in human breast carcinoma: a multiomic approach Cancer Inform [Internet] 19 117693512096969 Date accessed: 01/10/24 https://doi.org/10.1177/1176935120969696
24. Zhang YC, Zhao H, and Chen C, et al (2020) COX-2 gene rs689466 polymorphism is associated with increased risk of colorectal cancer among Caucasians: a meta-analysis World J Surg Oncol [Internet] 18 192 [https://wjso.biomedcentral.com/articles/10.1186/s12957-020-01957-x] Date accessed: 01/10/24
25. Mädge JC, Stallmach A, and Kleebusch L, et al (2022) Meta-analysis of aspirin-guided therapy of colorectal cancer J Cancer Res Clin Oncol [Internet] 148 1407–1417 Date accessed: 01/10/24 https://doi.org/10.1007/s00432-022-03942-1
26. Wu QB and Sun GP (2015) Expression of COX-2 and HER-2 in colorectal cancer and their correlation World J Gastroenterol [Internet] 21 6206 [http://www.wjgnet.com/1007-9327/full/v21/i20/6206.htm] Date accessed: 01/10/24 https://doi.org/10.3748/wjg.v21.i20.6206
27. Kosumi K, Hamada T, and Zhang S, et al (2019) Prognostic association of PTGS2 (COX-2) over-expression according to BRAF mutation status in colorectal cancer: results from two prospective cohorts and CALGB 89803 (Alliance) trial Eur J Cancer [Internet] 111 82–93 [https://linkinghub.elsevier.com/retrieve/pii/S0959804919300437] Date accessed: 01/10/24 https://doi.org/10.1016/j.ejca.2019.01.022
28. Mima K, Nishihara R, and Yang J, et al (2016) MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival Clin Cancer Res [Internet] 22 3841–3848 [https://aacrjournals.org/clincancerres/article/22/15/3841/79201/MicroRNA-MIR21-miR-21-and-PTGS2-Expression-in] Date accessed: 01/10/24 https://doi.org/10.1158/1078-0432.CCR-15-2173
29. Xie WL, Chipman JG, and Robertson DL, et al (1991) Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing Proc Natl Acad Sci [Internet] 88 2692–2696 Date accessed: 01/10/24 https://doi.org/10.1073/pnas.88.7.2692
30. Brown JR and DuBois RN (2005) COX-2: a molecular target for colorectal cancer prevention J Clin Oncol [Internet] 23 2840–2855 Date accessed: 01/10/24 https://doi.org/10.1200/JCO.2005.09.051
31. Wang D, Wang H, and Shi Q, et al (2004) Prostaglandin E2 promotes colorectal adenoma growth via transactivation of the nuclear peroxisome proliferator-activated receptor delta Cancer Cell 6(3) 285–295 https://doi.org/10.1016/j.ccr.2004.08.011
32. Williams CS, Sheng H, and Brockman JA, et al (2001) A cyclooxygenase-2 inhibitor (SC-58125) blocks growth of established human colon cancer xenografts Neoplasia [Internet] 3 428–436 [https://linkinghub.elsevier.com/retrieve/pii/S1476558601800567] Date accessed: 01/10/24 https://doi.org/10.1038/sj.neo.7900177
33. Peacock O, Lee AC, and Cameron F, et al (2024) Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer PLOS One [Internet] 9 e110267 [https://dx.plos.org/10.1371/journal.pone.0110267] Date accessed: 01/10/24
34. De Moraes E, Dar NA, and De Moura Gallo CV, et al (2007) Cross-talks between cyclooxygenase-2 and tumor suppressor protein p53: balancing life and death during inflammatory stress and carcinogenesis Int J Cancer [Internet] 121 929–937 Date accessed: 01/10/24 https://doi.org/10.1002/ijc.22899
35. Ghaleb AM and Yang VW (2017) Krüppel-like factor 4 (KLF4): what we currently know Gene [Internet] 611 27–37 [https://linkinghub.elsevier.com/retrieve/pii/S0378111917301142] Date accessed: 01/10/24 https://doi.org/10.1016/j.gene.2017.02.025
36. Hidalgo-Estévez AM, Stamatakis K, and Jiménez-Martínez M, et al (2020) Cyclooxygenase 2-regulated genes an alternative avenue to the development of new therapeutic drugs for colorectal cancer Front Pharmacol [Internet] 11 533 [https://www.frontiersin.org/article/10.3389/fphar.2020.00533/full] Date accessed: 01/10/24 https://doi.org/10.3389/fphar.2020.00533
37. Alinia S, Ahmadi S, and Mohammadi Z, et al (2024) Exploring the impact of stage and tumor site on colorectal cancer survival: Bayesian survival modeling Sci Rep [Internet] 14 4270 Date accessed: 01/10/24 https://doi.org/10.1038/s41598-024-54943-8
38. Monteleone NJ, Moore AE, and Iacona JR, et al (2019) miR-21-mediated regulation of 15-hydroxyprostaglandin dehydrogenase in colon cancer Sci Rep [Internet] 9 5405 Date accessed: 01/10/24 https://doi.org/10.1038/s41598-019-41862-2
39. Wei J, Zhang J, and Wang D, et al (2022) The COX-2–PGE2 pathway promotes tumor evasion in colorectal adenomas Cancer Prev Res (Phila Pa) [Internet] 15 285–296 [https://aacrjournals.org/cancerpreventionresearch/article/15/5/285/694474/The-COX-2-PGE2-Pathway-Promotes-Tumor-Evasion-in] Date accessed: 01/10/24 https://doi.org/10.1158/1940-6207.CAPR-21-0572
40. Bengi G, Keles D, and Topalak Ö, et al (2015) Expressions of TIMP-1, COX-2 and MMP-7 in ColonPolyp and colon cancer Euroasian J Hepato-Gastroenterol [Internet] 5 74–79 Date accessed: 01/10/24 https://doi.org/10.5005/jp-journals-10018-1138
41. Hall DCN and Benndorf RA (2022) Aspirin sensitivity of PIK3CA-mutated colorectal cancer: potential mechanisms revisited Cell Mol Life Sci [Internet] 79 393 Date accessed: 01/10/24 https://doi.org/10.1007/s00018-022-04430-y
42. Lu Y, Shi C, and Qiu S, et al (2016) Identification and validation of COX-2 as a co-target for overcoming cetuximab resistance in colorectal cancer cells Oncotarget [Internet] 7 64766–64777 [https://www.oncotarget.com/lookup/doi/10.18632/oncotarget.8649] Date accessed: 01/10/24
43. Lin JL, Lin JX, and Zheng CH, et al (2020) Relationship between aspirin use of esophageal, gastric and colorectal cancer patient survival: a meta-analysis BMC Cancer [Internet] 20 638 [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-020-07117-4] Date accessed: 01/10/24
44. Luo C, Cen S, and Ding G, et al (2019) Mucinous colorectal adenocarcinoma: clinical pathology and treatment options Cancer Commun [Internet] 39 1–13 Date accessed: 01/10/24 https://doi.org/10.1186/s40880-019-0361-0
45. Verhulst J, Ferdinande L, and Demetter P, et al (2012) Mucinous subtype as prognostic factor in colorectal cancer: a systematic review and meta-analysis J Clin Pathol [Internet] 65 381–388 Date accessed: 01/10/24 https://doi.org/10.1136/jclinpath-2011-200340
46. Dai D, Zhou B, and Zhong Y, et al (2019) Survival of patients with resected primary colorectal mucinous adenocarcinoma: a competing risk nomogram analysis Oncol Lett [Internet] 18(6) 6594–6604 [http://www.spandidos-publications.com/10.3892/ol.2019.11024] Date accessed: 01/10/24
47. Wendum D, Masliah J, and Trugnan G, et al (2004) Cyclooxygenase-2 and its role in colorectal cancer development Virchows Arch [Internet] 445 327–333 Date accessed: 01/10/24 https://doi.org/10.1007/s00428-004-1105-2